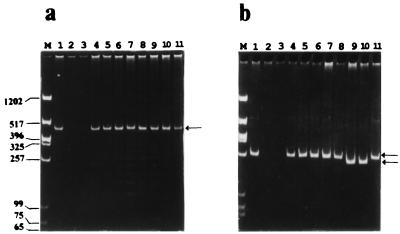
FIG. 3.
PCR analysis of RS I (a) and RS II (b) regions. PCRs were carried out (95°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 30 cycles), and the PCR products were run on 5% acrylamide gels. P1 and P2 were used to amplify RS I; P3 and P4 were used to amplify RS II. Genomic DNA isolated from the different yeast strains by the glass bead method (7) was used as the template for the PCRs. For marker DNA (lanes M) we used a HinfI/XbaI digest of pBSK(−). Lanes: 1, S. pombe ABP13 h− ade6-210 ura4-D18 (obtained from Fission Yeast Course); 2, S. japonicus var. versatilis (obtained from Fission Yeast Course); 3, S. octosporus (obtained from Fission Yeast Course); 4, S. pombe KN1 h− ade6-216 ura4-D18 leu1-32 pmd1::ura4+ (13); 5, S. pombe ABP 320 h+ arg3-D4 ura4-D18 (21); 6, S. pombe GP 25 h− his5-303 lys1-131 (3); 7, S. pombe h90 swi4-1 NCYC 1992; 8, S. pombe NCYC 1990; 9, S. pombe var. malidevorans NCYC 2387; 10, S. malidevorans NCYC 683; and 11, plasmid pCPY1 containing the original cpy1+ clone that was isolated in this laboratory (1). Arrows indicate the repeat regions. Molecular sizes of marker DNA are given (in base pairs) to the left of panel a.