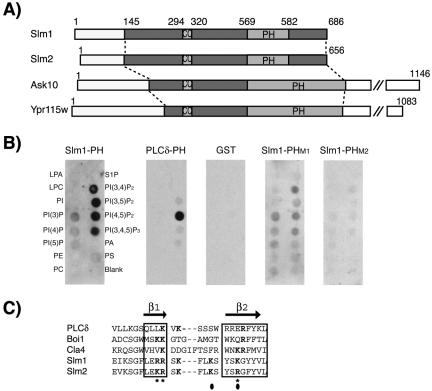
Figure 1.
Domain organization of Slm1 and Slm2 and lipid binding specificity. (A) Schematic diagram of Slm1, Slm2, Ask10, and Ypr115w structures showing the locations of PH (PH) and coiled-coil (CC) domains, respectively. Light and dark shaded boxes indicate the regions conserved among all four proteins. (B) Nitrocellulose-immobilized phospholipids (PIP Strip) were incubated with recombinant fusion proteins of GST to the C terminus of Slm1 (amino acids 441–686) containing either the wild-type PH domain (Slm1-PH) or the mutant variants PHM1, and PHM2. GST alone or a GST fusion to the PH domain of PLCδ, which specifically binds PtdIns(4,5)P2 in vitro were used as controls. Bound proteins were visualized by Western blot analysis with anti-GST antibodies. Spots contained lysophosphatidic acid (LPA), lysophosphacholine (LPC), PtdIns (PI), PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine-1-phosphate (S1P), PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, PtdIns(3,4,5)P3, phosphatic acid (PA), and phosphatidylserine (PS). (C) Sequence alignment of the β1/β2 regions of Slm1 and Slm2 PH domains with related PH domains from PLCδ, Boi1, and Cla4. Secondary structure is indicated above the sequence alignment. Bold letters indicate residues required for interaction with phosphoinositide ligands. Asterisks and closed circles indicate amino acid residues targeted for mutagenesis in SlmM1 and SlmM2 PH domain mutants, respectively.