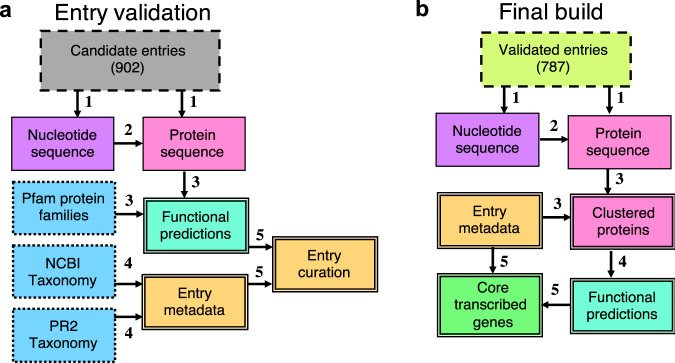
Fig. 1.
Diagrammatic overview of MarFERReT validation and build processes. Boxes represent the data sets involved in building MarFERReT and the border style indicates the data type: external sequence inputs (dashed line), external taxonomic and functional annotation resources (dotted lines), internal data products (single solid line) and output MarFERReT data products (double lines). Arrows indicate processes. (a) Candidate entry and NCBI taxID validation: (1) Candidate entries were identified from primary data sources and downloaded as nucleotide and protein reference sequences; (2) six-frame translation301 and frame-selection of nucleotide sequences into protein sequences; (3) functional annotation of protein sequences with Pfam292 protein families using HMMER 3.3302; (4) curation of NCBI Taxonomy293 IDs (taxIDs) for MarFERReT candidate entries and additional incorporation of matched IDs and classification from the PR2 Taxonomy ecosystem294,295; (5) candidate entries are assessed with evidence from external studies and by taxonomic analysis of ribosomal protein sequences for potential cross-contamination. Validated entries accepted for the quality-controlled build are recorded in the entry metadata. (b) Quality-controlled MarFERReT build with validated entries. For the set of 800 validated entries, the same methods used in 1a were used for (1) aggregating nucleotide and protein data and (2) translating nucleotide to protein sequences; (3) intra-taxa clustering at the strain or species level: protein sequence data sharing the same NCBI taxIDs are pooled together and clustered307 at 99% identity using updated taxIDs contained in the metadata; (4) Final Pfam annotation of the clustered protein sequences; (5) identification of core transcribed genes from functional annotations of transcriptome-derived entries.