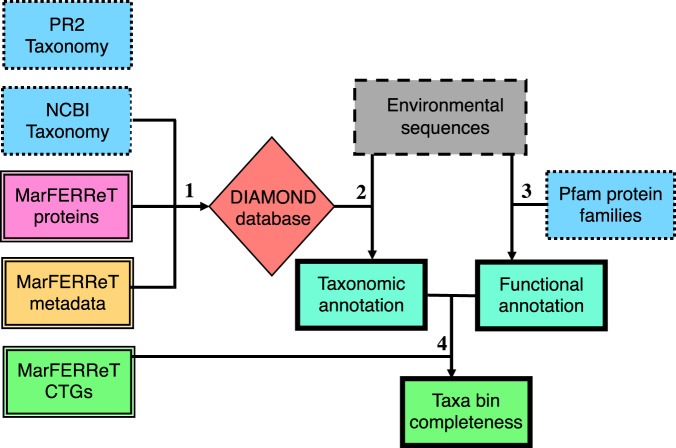
Fig. 4.
Schematic of case study 1 and 2 use of MarFERReT for annotation of environmental metatranscriptomes. Example workflow showing how MarFERReT data products can be used to annotate unknown assembled sequences and assess taxonomic bins. Boxes indicate datasets; box borders indicate environmental contig sequence data (dashed line), taxonomic and functional annotation from external resources (dotted lines), MarFERReT data products (double lines) and taxonomic and functional annotation results (bold lines). The red diamond indicates a user-constructed DIAMOND306 database for lowest common ancestor determination; prior to this step the user could combine MarFERReT with other libraries for expanded taxonomic coverage as shown in case study 1. Arrows represent processes: (1) construction of DIAMOND306 database using MarFERReT proteins and data, and taxonomy files from NCBI Taxonomy; matching PR2 Taxonomy294,295 identifiers and classifications are also provided in the metadata for alternative classification approaches. (2) Taxonomic annotation of environmental contigs using a DIAMOND306 database built from MarFERReT proteins; (3) Functional annotation of environmental contigs with HMMER302 3.3 on Pfam292 protein family hmm profiles; (4) Completeness assessment of taxonomically- and functionally-annotated metatranscriptome bins using MarFERReT core transcribed genes.