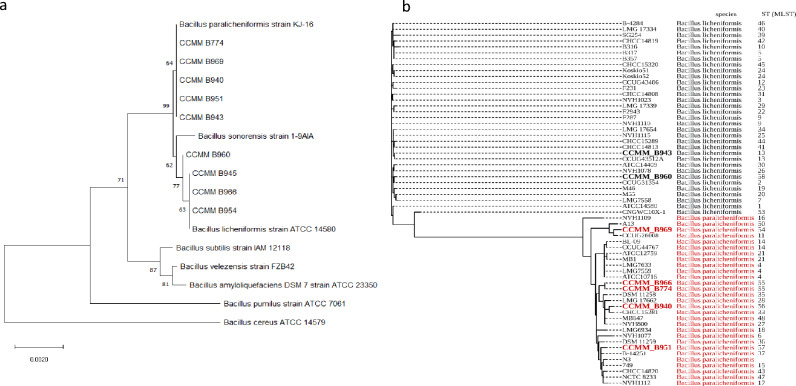
(a) Neighbor-joining phylogenetic tree based on 16S rRNA sequences of Bacillus isolates. The phylogenetic tree was inferred using the Neighbor-Joining method62. The percentage of replicate trees in which associated taxa clustered in the bootstrap test (1000 replicates) is shown next to the branches (bootstrap values ≥ 60). Evolutionary analyses were conducted in MEGA X63. The bar indicates 0.0020 substitution per site. rRNA gene sequences are presented with the corresponding species name and strain, or with the CCMM reference. (b) Multi Locus Sequence Typing (MLST) analysis of B. licheniformis and B. paralicheniformis strains. The phylogenetic tree was generated in iTOL v4 integrated into the PubMLST database, based on the internal fragments of six housekeeping genes. Seven Bacillus CCMM strains were submitted to the database. Bacillus isolates are presented with the corresponding species and strain names.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.