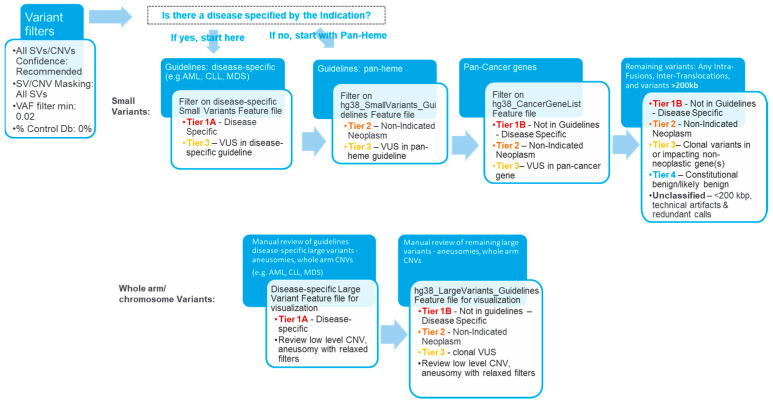
Figure 1.
Overview of the analysis and interpretation workflow. The curation and classification process consists of four steps: (1) Disease-specific: Several organizations, such as the World Health Organization (WHO), the National Health Service (NHS), and the National Comprehensive Cancer Network (NCCN), have published guidelines for assessing clinically relevant genomic regions for hematological malignancies. The analysts apply a filter to select for variants overlapping with known loci associated with the disease of the sample (e.g., AML), then classify the selected SVs as Tier 1A or 3. (2) Pan-hematological cancers: The analyst removes the first filter and applies another filter to select for SVs seen in hematological cancers and classifies them as Tier 2 or 3. (3) Pan-cancer: SVs overlapping other cancer-associated genes are selected and classified as Tier 1B, 2, 3, or 4. (4) Remaining variants: All remaining fusion SVs or SVs >200 kbp are classified as Tier 1B, 2, 3, or 4.