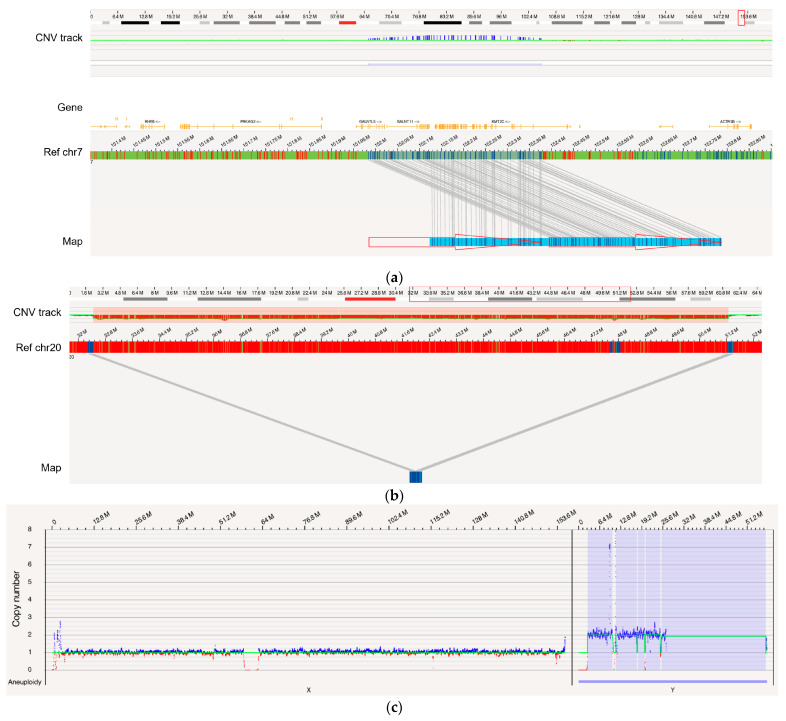
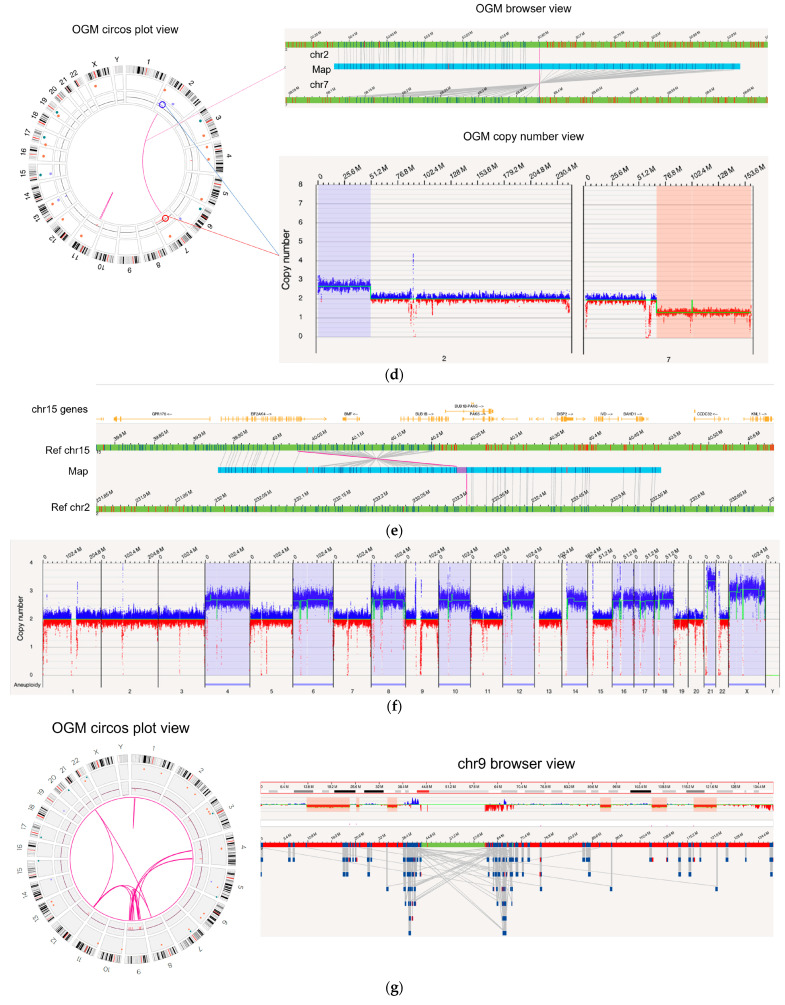
Figure 3.
Examples of several types of SVs examined in comparison with SOC. (a) A 392.8 kbp tandem duplication occurred in the genome of BNGOHM-0000191, an MDS sample, with a VAF of 1.0. The two red arrows indicate the two copies of the duplicon on the assembled map. (b) A 18.8 Mbp deletion with a VAF of 0.44 was observed in MPN genome BNGOHM-0000176. Note that both the coverage-based copy-number algorithm and the map-alignment-based algorithm captured the deletion. (c) In ALL sample BNGOHM-0000172, a whole-genome duplication of chromosome Y was observed. (d) OGM captured an unbalanced t(2;7) in the MDS genome of sample BNGOHM-0000138. The blue and red lines indicate the translocation break points, which coInc.ide with a copy-number gain on chr2 and a loss on chr7. The VAF of the translocation was 0.28. (e) A 168.9 kbp inversion adjacent to a t(2;15) translocation was captured by a long genome map. The events occurred at a VAF of 0.27, and the BUB1B gene was interrupted by the inversion break point. (f) A hyperdiploid genome in ALL genome BNGOHM-0000333 as seen in OGM’s whole-genome coverage view. (g) OGM resolved the structure of a complex ALL genome of sample BNGOHM-0000243.