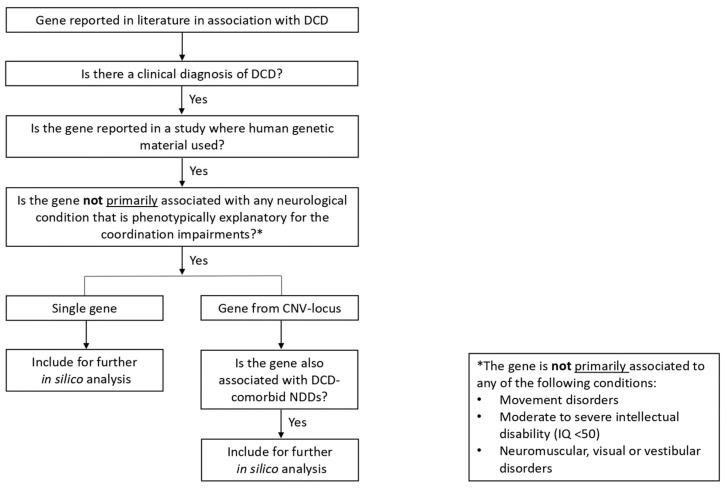
Figure 1.
Gene inclusion criteria for the present study. Flowchart depicting the process of gene inclusion based on the inclusion criteria that we defined according to the official guidelines [2,3]. First, we examined whether there was a clinical diagnosis of DCD according to the DSM-4 or DSM-5 criteria. We only included genes reported in patients with a clinical diagnosis of DCD. Genes reported in patients who were suspected of DCD or had a similar phenotype but no clinical diagnosis, were not included. Second, we screened for genetic variants that were reported in studies using human genetic material. Candidate genes from mice studies were therefore not included. Genetic variants were reported as either single nucleotide polymorphism (SNP) or copy-number variations (CNV) in a genetic locus. Third, we investigated the disease associations of these genes. According to the official diagnostic criteria, we only included genes that were not primarily associated with any of the following conditions: movement disorders, moderate to severe ID (defined in the ICD-10 as an IQ of <50), or neuromuscular, visual or vestibular disorders. If all of the abovementioned criteria were met, then we included the gene for further in silico analysis. For genes residing within a CNV-locus, we further checked whether the gene was previously associated with DCD-comorbid neurodevelopmental disorders (NDDs), such as attention deficit/hyperactivity disorder or autism-spectrum disorder. If that was the case, then the gene was included for further in silico analysis. DCD = Developmental Coordination Disorder; SNP = single nucleotide polymorphism; CNV = copy-number variation; NDD = neurodevelopmental disorder.