Fig. 4.
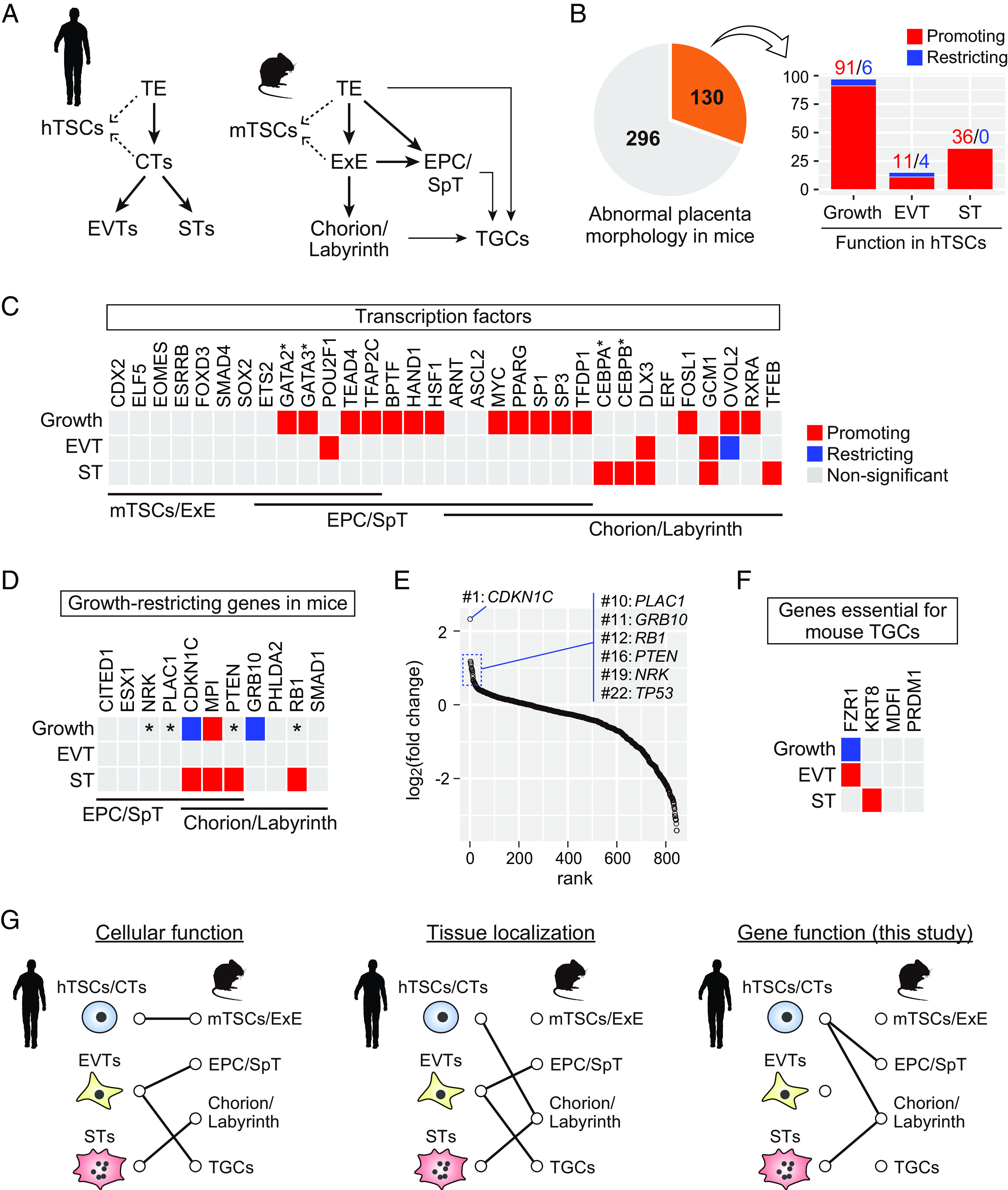
Comparison of gene functions between hTSCs and mouse placenta. (A) Schematic representation of human and mouse trophoblast development. (B) Identification of genes required in both hTSCs and mouse placenta. Among the 426 genes implicated in abnormal placenta morphology in mice, 130 genes were classified as essential for hTSC growth or differentiation. Genes that promote hTSC growth or differentiation are shown in red, and those that restrict hTSC growth or differentiation are in blue. (C) Analysis of TFs involved in the development of mTSCs/ExE, EPC/SpT, or chorion/labyrinth in mice. Genes promoting hTSC growth or differentiation are shown in red, genes restricting hTSC growth or differentiation are in blue, and those without significant effect are in gray. *Note that Gata2/3 (30) and Cebpa/b (31) are functionally redundant in the mouse placenta. (D) Analysis of genes whose KO leads to an enlarged placenta in mice. Genes are color-coded as in (C). *Note that although not statistically significant (FDR > 0.05), these genes were highly ranked as negative growth regulators as shown in (E). (E) Results of CRISPR screening for hTSC growth regulators. Genes are ordered by their log2(fold change) values. Growth-restricting genes have high log2(fold change) values. (F) Analysis of genes essential for TGC differentiation in mice. Genes were color-coded as in (C). See also SI Appendix, Table S1 for a detailed listing of placental phenotypes of relevant KO mice. (G) (Left) Cellular function-based prediction. hTSCs/CTs share similarities with mTSC/ExE as both are proliferative and can give rise to differentiated trophoblasts. EVTs invade into the maternal uterus and remodel the spiral arteries. These functions are mediated by EPC/SpT and TGCs in mice. STs and the chorion/labyrinth mediate gas and nutrient exchange. (Middle) Tissue localization-based prediction. Human placental villi, which contain CTs and STs, are structurally similar to the chorion/labyrinth in mice. EVTs migrate out from placental villi and come into direct contact with uterine cells. Glycogen trophoblasts in the EPC/SpT and TGCs directly interact with uterine cells in the mouse placenta. (Right) Gene function–based prediction. Our CRISPR screening results suggest that hTSCs may be most similar to the EPC/SpT and chorion/labyrinth (possibly progenitor cells in these tissues), whereas STs are analogous to the chorion/labyrinth (possibly ST layer I and II). Alternatively, the exact counterpart of EVTs in mice could not be clearly determined.
