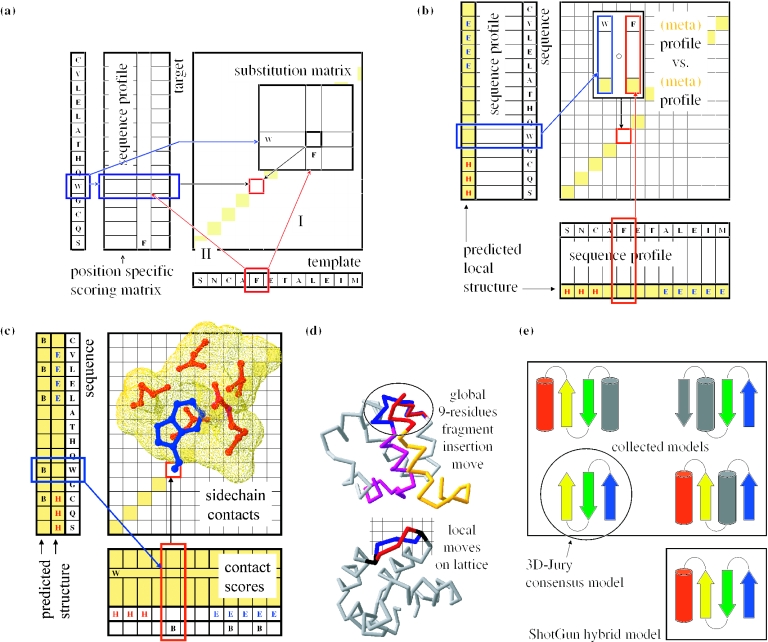
Figure 2.
Protein structure prediction methods. (a) Sequence–sequence, profile–sequence, sequence–profile comparison methods represent a traditional evolutionary-based approach to predict structures of proteins. The simplest method (I) aligns the sequence of the target with the sequence of the template using a substitution matrix. More sensitive methods (II) define scores for aligning different amino acids separately for each position of the target sequence (PSI-BLAST) or the template sequence (RPS-BLAST). The scores are taken from the analysis of sequence variability in multiple alignments of the corresponding sequence families. Such position-specific scores are also called profiles. They are similar in format to the representation of sequence families used by prediction methods based on HMMs. (b) Profile–profile comparison methods utilize the profiles generated by the above mentioned sequence alignment methods. Instead of a lookup of a substitution score, they compare two vectors with each other when building the dynamic programming matrix used to draw the alignment. The comparison is usually conducted by calculating a dot product of the two positional vectors (as shown in the figure) or by multiplying one vector times a substitution matrix time the other vector. Depending on the choice of the comparison function the vectors are often rescaled before the operation. The sequence variability vectors are sometimes also augmented with meta information, such as predicted secondary structure as indicated in the figure. (c) Threading or hybrid methods utilize the structure of the template protein in the comparison function. The position-specific alignment scores are computed for the template protein by replacing the side-chain of a residue with side-chains of all possible amino acids and by calculating the resulting substitution scores using statistically derived contact potentials. In addition, factors such as matching of predicted and observed secondary structure or burial preferences are also taken into account when aligning two positions. Most threading methods use frozen approximation where the sequence is threaded through the template structure and contacts are calculated between the target side-chain and side-chains of the residues of the template. In the much slower, defrosted threading template side-chains are replaced with side-chains of the target according to the alignment before calculating the contact scores. (d) Ab initio methods represent a physical approach to predict the structure of the target protein. The methods are based on an energy function, which estimates the conformational energy of the chain of the modeled protein. The energy can be calculated in a similar fashion as in the threading methods, i.e. utilizing contact potentials. The advantage of ab initio is that the database of folds does not constrain the set of possible results and theoretically any conformation can be generated and tested. Ab initio methods differ in employed energy functions and in the way conformational modifications are generated. Most common methods employ fragment insertion techniques or constrain the move set by placing the molecule on a lattice. (e) Meta predictors represent statistical approaches to improve the accuracy of protein structure predictions. Simple meta predictors collect models from prediction servers, compare the models and select the one, which is most similar to other models. The consensus model corresponds to a model selected from the collected set and represents the final prediction. More advanced meta predictors are able to modify the set of collected models either by filing missing parts with ab initio or loop modeling or by creating hybrid models from segments of structures collected from prediction servers. Hybrid models have a higher chance to provide a more complete model but are sometimes unphysical in terms of chain connectivity.