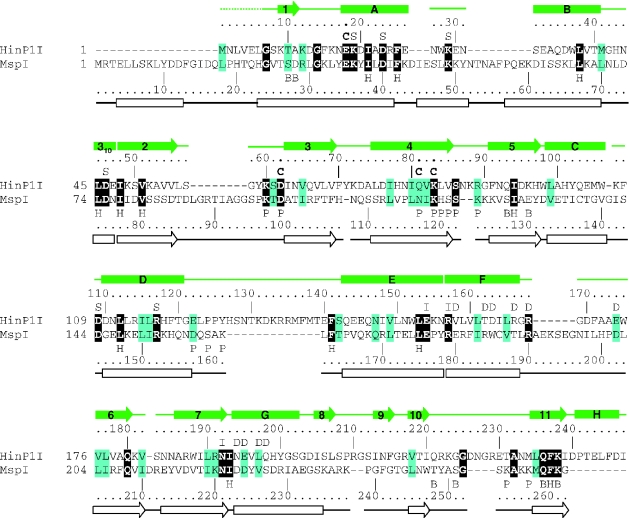
Figure 3.
Structure-based sequence alignment of HinP1I and MspI. The residue number and secondary structural elements of HinP1I and MspI are shown, respectively, above and below the aligned sequences. The amino acids highlighted are invariant (white letter against black background) and conserved (black against cyan). The letters immediately above or below the sequences indicate the structural and suggested functional roles of the corresponding residues: ‘B’ indicates DNA base interaction, ‘C’ indicates catalysis, ‘D’ indicates dimer interface, ‘H’ indicates hydrophobic core, ‘I’ indicates intra-molecule interaction, ‘P’ indicates DNA phosphate interaction and ‘S’ indicates conserved surface residues (whose locations are shown in Figure 2B). The protein–DNA interactions involving main chain atoms are not indicated.