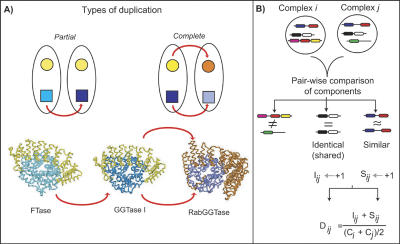
Figure 1.
Detection of duplicated proteins complexes. (A) Schematic illustrating two types of duplication of protein complexes. In the top panel on the left, we show a partial duplication in which one of the proteins in the complex has duplicated, and the other protein is part of both resulting complexes. On the right, a duplication of both components of the complex is illustrated, which we term `complete duplication' of a complex. In the bottom panel, we give an example of yeast protein complexes that follow the two duplication scenarios. The heterodimeric complexes farnesyl transferase (FTase), genranylgeranyl transferase I (GGTase I) illustrate a partial duplication. They share one subunit (α subunit, shown in yellow), but each has a distinct β subunit, coded by paralogous genes (shown in shades of blue) (Casey and Seabra 1996). GGTase I and Rab Geranylgeranyl Transferase (RabGGTase) illustrate a complete duplication. Paralogous genes code for the α and β subunits of both complexes (Casey and Seabra 1996). (B) Schematic representation of the method used to identify duplicated complexes. Each pair of complexes is compared in terms of their components to ascertain whether they are homologous, having evolved by partial or complete duplication. Any components that are shared, in other words that are identical proteins, are counted, as represented by the variable I. Any components that are homologous according to their domains as assigned by the Pfam or SUPERFAMILY databases, or by sequence similarity, are counted as similar components, represented by the variable S. For each pair of complexes, a score based on S, I, and the sizes of the two complexes (C) is calculated. If the score exceeds a certain threshold, we consider the two as duplicate (or homologous) complexes. If the two complexes have similar as well as identical components, they have evolved by partial duplication, and if only similar components are present, they have evolved by complete duplication.