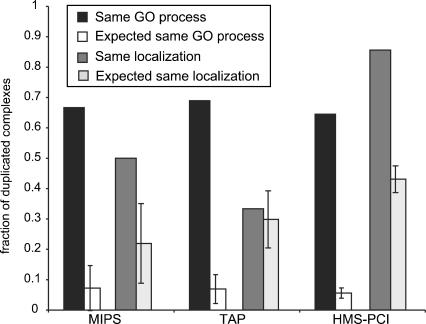
Figure 3.
Functional consequences of complex duplication. Proportion of pairs of duplicated complexes belonging to the same GO biological process and to the same subcellular localization, compared with 10,000 random samplings of equal size of the data sets. In all cases pairs of duplicated complexes are significantly more likely to have the same functional assignment than could be expected by chance (zMIPS = 8.0, zTAP = 13.1, zHMS-PCI = 35.0, in all cases P ≤ 10–4). For similar subcellular localization, the proportion of complexes is significantly larger than could be expected by chance (zMIPS = 2.15 P ≤ 10–2, zHMS-PCI = 9.6 P ≤ 10–4) in both the MIPS and HMS-PCI data sets, and the TAP data set does not show any significant deviation from the random expectation. In an independent experiment controlling for shared proteins in duplicated complexes, we observed that in all data sets, and for both classification schemes, the proportion of homologous complexes with the same classification is significantly higher than expected by chance (P < 10–4).