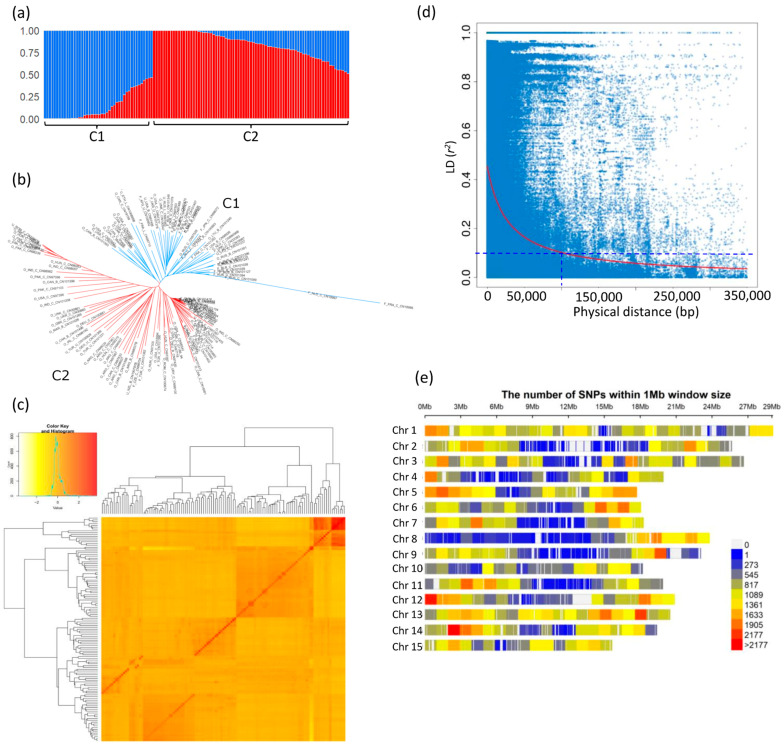
Figure 2.
(a) Model-based population structure of 123 flax accessions belonging to two clusters predefined by the STRUCTURE software. (b) Neighbor-joining (NJ) tree of 123 flax accessions based on the 272,944 SNPs. (c) Kinship relationships among the 123 flax accessions of the collection. (d) Genome-wide linkage disequilibrium (LD) decay of r2 values (red line), against physical distance (bp) in flax. The dashed blue line indicates the cutoff value (r2 = 0.1) used to determine the genome-wide LD block size. (e) Distribution of 272,944 SNPs used for GWAS analysis across the 15 Linum usitatissimum chromosomes. C1: cluster 1; C2: cluster 2.