Fig. 6.
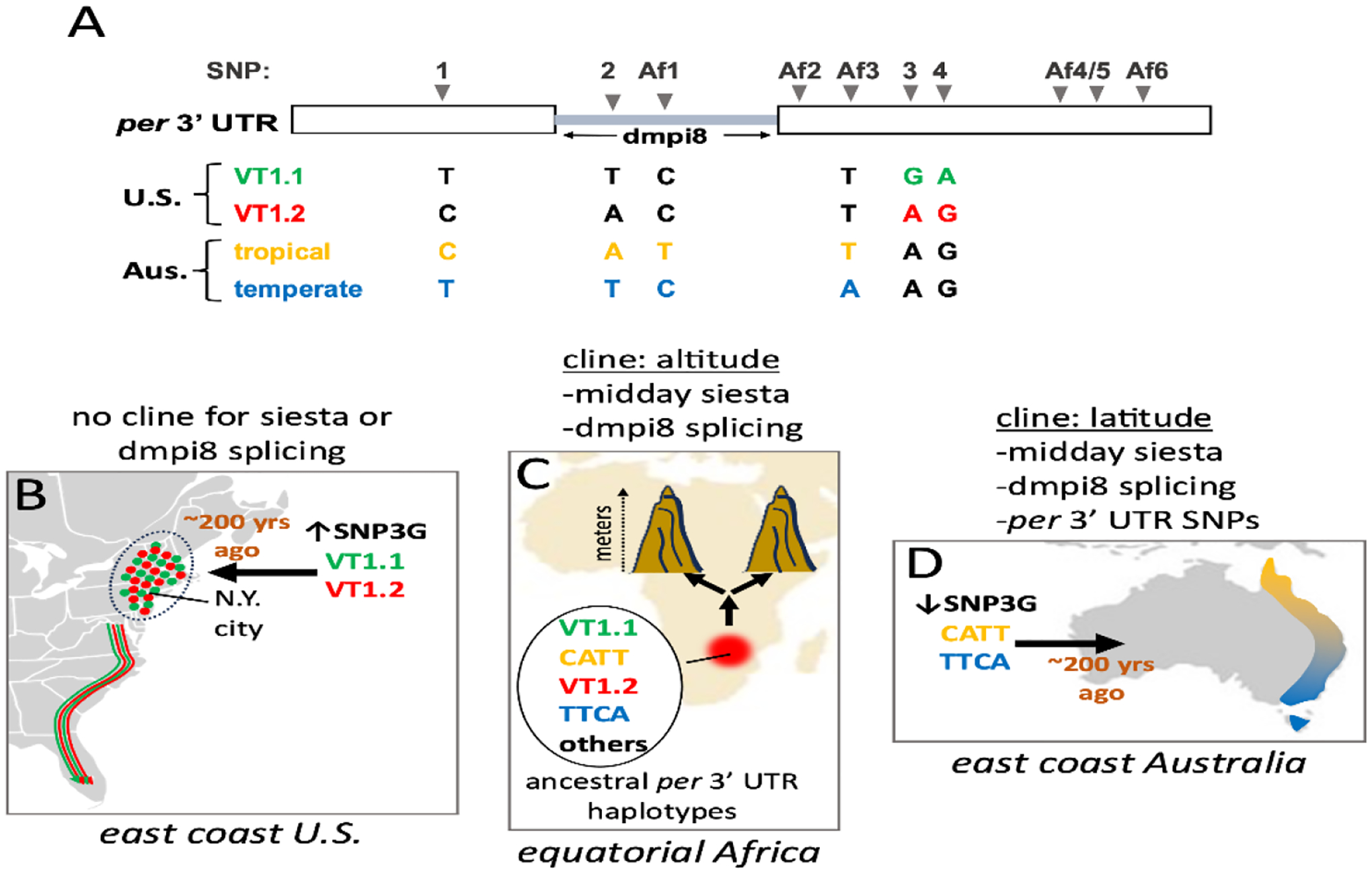
Summary of clinal relationships between midday siesta, dimpi8 splicing efficiency and per 3’ UTR SNPs. (A) Schematic of of the major SNPs in the per 3’ UTR found in natural populations of D. melanogaster that we sampled. (B, C, D) While all per3’ UTR SNPs identified to date are found in Africa, presumably also in ancestral populations (C), it is possible that the founding populations for North America (B) and Australia (D) differed in per3’ UTR haplotypes, perhaps explaining why latitudinal clines in dmpi8 splicing and midday siesta are observed in Australia but not the northeastern United States.
