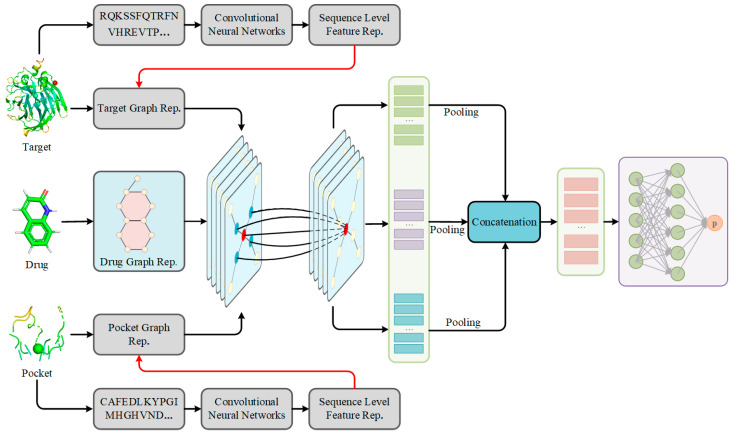
Figure 5.
The architecture of S2DTA. S2DTA is an advanced model designed to predict drug–target affinity (DTA) by leveraging sequence features of drug SMILES, targets, and pockets, and their corresponding structural features. This model comprises four essential modules: (1) The data input module was responsible for representing the sequence and structural data of drugs, targets, and pockets. (2) Within the sequence learning module, a 1D-CNN layer was employed to extract semantic features from both targets and pockets, based on their sequence data. (3) In the structure learning module, three independent Graph Convolutional Networks (GCNs) were employed to extract high-level features from the vertices present in the graph-based representation of drugs, targets, and pockets. (4) The feature fusion module encompasses a two-layer fully connected (FC) network, which played a pivotal role in predicting DTA by integrating the extracted features.