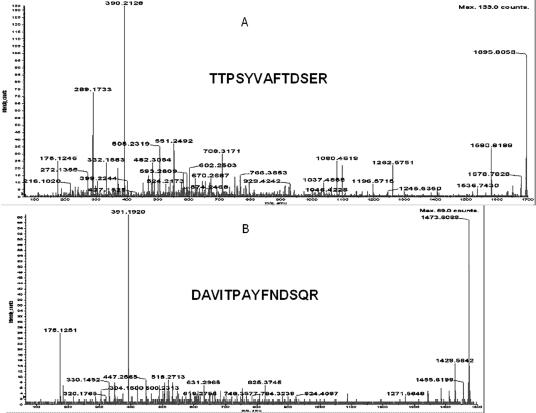
Fig 3.
Microsequencing of the proteins which were identified as heat shock protein (Hsp)70 (A) and Hsp68 (B) using trypsin fingerprinting and database search. The determined sequences are presented in bold. Protein identification was done by trypsin fingerprinting using surface-enhanced laser desorption ionization–time-of-flight–mass spectrometry (SELDI-TOF-MS) (Ciphergen Biosystems Inc.) followed by NCBI database search using the Profound search engine and confirmed by SELDI-TOF-MS MS (Ciphergen Biosystems Inc.) analysis. All proteins used in these experiments were obtained as gel slices after two-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis stained with Coomassie Blue (G-250). “No protein” control gel slices were included into each experiment. Gel slices were processed exactly as recommended in Ciphergen product insert to Protein ID kit up to gel drying step (Stage B, point 7). The following changes were introduced to the subsequent steps of trypsin in-gel digestion and peptide recovery: (1) trypsin (Roche Applied Science, Basel, Switzerland) was dissolved in water to the final concentration of 1 μg/μL; (2) dried and crushed gel slices were resuspended in 50 μL of 50 mM ammonium bicarbonate buffer, pH 8.0 (freshly prepared) containing 12.5 ng/μL of trypsin and digested for 16 hours at 37°C; (3) supernatants were collected and freeze-dried in SpeedVac for 20 minutes; (4) peptides were dissolved in 10 μL of H2O, 3 μL was applied per spot of H4 ProteinChip® array (Ciphergen Biosystems Inc.), and air dried; (5) each spot of the array was washed with 5 μL of 5% acetonitrile in water 2 times and air dried; and (6) 1 μL of 20% CHCA in 50% acetonitrile–0.5% trifluoroacetic acid (TFA) was applied per spot for Protein Chip Array Reader (PBSII) SELDI-TOF (Ciphergen Biosystems Inc.) analysis. One microliter of saturated cyano-4-hydroxy-cinnamic acid (CHCA) in the same solution was added per spot of array followed by microsequencing using PCI-1000 interface (Ciphergen Biosystems Inc.) to QStar I (ABI Sciex) tandem MS instrument. PBSII was calibrated externally with All-in-1 peptide standard (Ciphergen Biosystems Inc.) spot by spot, and calibration equations for each spot were saved separately. After calibration, the same chip was reread automatically twice and calibration equations for each spot were applied, correspondingly, each time. Recalibration and change of the equation were introduced, if needed. The experimental chip was read in automatic mode using laser intensity 190 and sensitivity 9. Saved calibration equations were applied to each spot, correspondingly. All peaks with signal to noise ratio above 2.5 were labeled automatically and peptides masses were used for NCBI database search using Profound (old or new version) for trypsin fingerprinting identification. All labeled peak mass values were included into search using automatic introduction by Ciphergen ProteinChip™ software 3.0. Search was done using the following parameters: Cys modified by acrylamide, mass error 1.0 Da, and number of miscleavages −1. The same database was searched using Mascot search engine and masses of fragmented parent ions produced by PCI-1000-QStar analysis to confirm the identification done by trypsin fingerprinting analysis.