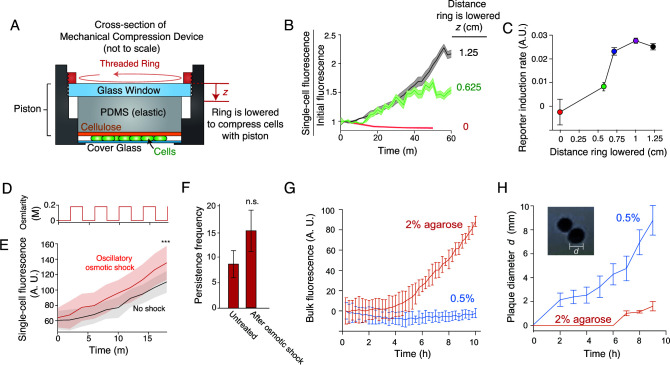
Fig 4.
Several forms of mechanical stimulation cause persistence. (A) A novel mechanical compression device in which cells are compressed with an elastic piston by sandwiching the piston between a cover glass and a threaded aluminum ring that can be lowered (by screwing it into the cover glass holder) by defined amounts. (B) Population-averaged single-cell fluorescence versus time and degree of compression in the device shown in (A), for E. coli cells harboring a plasmid-based transcriptional reporter for Rcs activity. Confidence intervals indicate ±1 SD across cells at each respective height. (C) Population-averaged single-cell induction rate of the reporter for Rcs activity versus degree of compression. n = 10, 181, 459, 790, 534 cells at each respective compression level. Error bars indicate ±1 SEM. (D) Medium osmolarity versus time during a 200-mM oscillatory osmotic shock with a 4-minute period. (E) Population-averaged single-cell fluorescence versus time for untreated (non-shocked) E. coli cells harboring a plasmid-based transcriptional reporter for Rcs activity and for the same strain undergoing an oscillatory osmotic shock as shown in (D). Confidence intervals indicate ±1 SD across n = 35 and 15 cells for the two conditions, respectively. ***: P < 0.0001. (F) Mean persistence frequency for untreated (non-shocked) E. coli and for the same strain after having undergone an oscillatory osmotic shock as shown in (D). Error bars indicate ±1 SEM. (G) Mean bulk fluorescence versus time of E. coli cells harboring a plasmid-based transcriptional reporter for Rcs activity that are suspended in solid media containing different concentrations of agarose. Error bars indicate ±1 SD across three technical replicates. (H) Mean plaque size versus time when a single bacteriophage solution was diluted into suspensions of bacteria in solid media containing 2% and 0.5% agarose. Error bars indicate ±1 SD across three technical replicates. (Inset) Micrograph of two bacteriophage plaques.