Figure 3. Functional annotation of the chromosome 3 SNP locus.
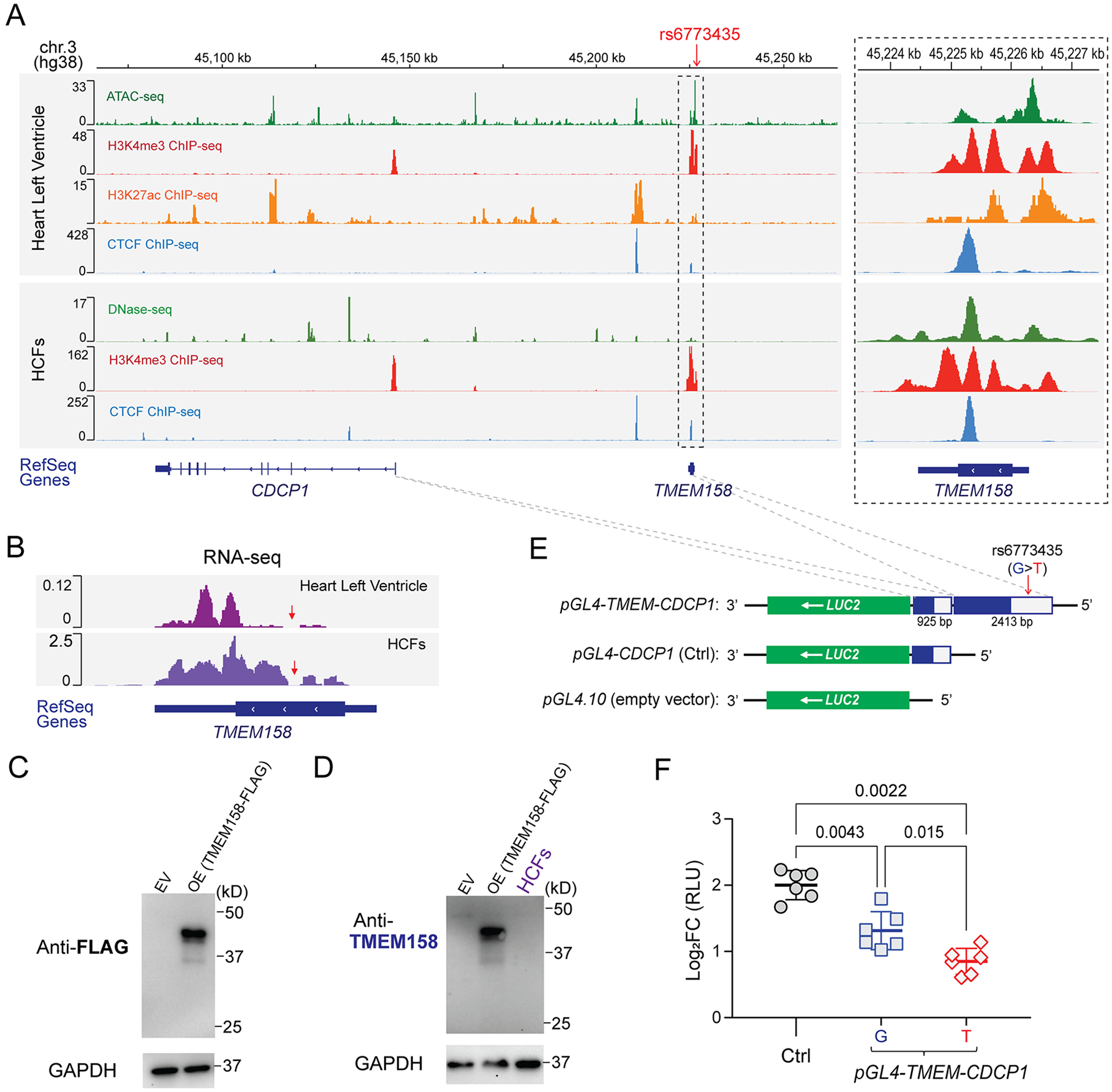
(A) Visualization of the ENCODE epigenomic dataset in the interactive genome viewer (IGV). Panels (from top to bottom) show the physical position of the rs6773435 SNP locus on chromosome 3 based on human genome assembly (hg38); ATAC-seq to assess genome-wide chromatin accessibility, ChIP-seq for H3K4me3 (promoter marker), for H3K27ac (enhancer and promoter marker), and for CTCF (chromatin looping marker) that were generated using human left ventricle tissues; DNase-seq to assess genome-wide chromatin accessibility, ChIP-seq for H3K4me3, and for CTCF that were generated using HCFs. (ATAC-seq and H3K27ac ChIP-seq data are not available for the HCFs). Genes were annotated by the NCBI human genome RefSeq. Both the TMEM158 and CDCP1 genes are transcribed from the negative DNA strand. The dashed box on the right panel is a “zoom-in” of the SNP locus. Sequencing peaks are map to the SNP locus, which indicates that the locus is likely a transcriptional activating site. (B) RNA-seq reads generated from human heart left ventricular tissue and HCFs which map to the TMEM158 gene annotated by RefSeq. The TMEM158 open reading frame (ORF) and untranslated region (UTR) were depicted by thick and thin lines, respectively. Red arrows indicate a region of the TMEM158 ORF where no RNA-seq reads were mapped. (C) Western blots using an anti-FLAG antibody for the putative TMEM158-FLAG fusion protein after overexpression (OE) in HEK-293T cells. Empty vector (EV) control sample was obtained from cells transfected with pCMV-Entry plasmid. (D) Western blot using anti-TMEM158 antibody for endogenous TMEM158 protein in HCFs. Overexpressed TMEM158-FLAG fusion protein sample, as same as which is blotted in (C) with anti-FLAG antibody, was blotted with the anti-TMEM158 antibody, which successfully detects overexpressed TMEM158-FLAG fusion protein. However, no endogenous TMEM158 protein was detected in protein lysates from HCFs. (E) Construction of reporter gene plasmids. The pGL4.10 plasmid which includes a LUC2 reporter gene was used as the backbone construct and served as an empty vector control. The CDCP1 promoter region (925 bp) was cloned into the 5′-end of the LUC2 reporter gene and was used as positive control plasmid (pGL4-CDCP1) for CDCP1 transcriptional activity. To test the effect of the rs6773435 SNP locus on CDCP1 transcription, DNA fragments that included the rs6773435 SNP/TMEM158 locus (2413 bp) were cloned into the 5’-end of the pGL4-CDCP1 (pGL4-TMEM-CDCP1). The pGL4-TMEM-CDCP1 plasmid containing the rs6773435 SNP “G” allele was compared with that containing the “T” allele. (F) Comparison of luciferase activities in HCFs transfected with pGL4.10 (empty vector), pGL4-CDCP1 (Ctrl), pGL4-TMEM-CDCP1 “G” and pGL4-TMEM-CDCP1 “T”. Data are log2 fold change (FC) in relative light units (RLUs) when compared to pGL4.10 alone (empty vector) from biological replicates (n=6) showing as mean values ± SD. Mann-Whitney test was used to calculate the presented p-values.
