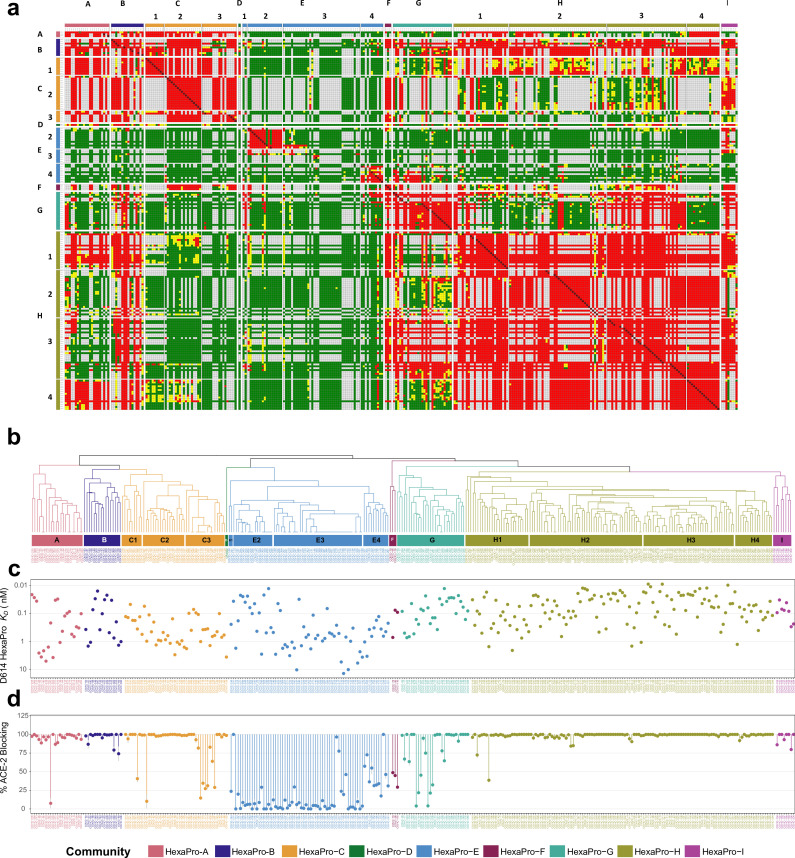
Fig 1.
Epitope binning on HexaPro separates the CoVIC panel into nine communities, with distinct completion profile, affinity range, and ACE-2 blocking ability. (a) Competition map of HexaPro binning communities from combining the results of two binning assays. Each colored square represents the categorization of normalized response for the specific pair of CoVIC constructs: red indicates competition (<0.5); green indicates sandwiching (>0.7); yellow indicates intermediate response (≥0.5 and ≤0.7); white indicates competition pairs where the data were not measured. The CoVIC IDs on the columns and rows correspond to the CoVIC IDs of the ligands (on chip surface) and the analytes (immune complexed with HexaPro), respectively. (b) Dendrogram derived from the competition map. The naming for each community and the corresponding color are shown in the legend (e.g., HexaPro-A, HexaPro-B). Sub-clusters of HexaPro-C, -E, and -H are indicated in panels a and b. (c) Binding KD value for D614 HexaPro separated by HexaPro binning community. (d) Percent blocking of ACE-2 to RBD binding separated by HexaPro binning community, with error bars shown as gray vertical lines.