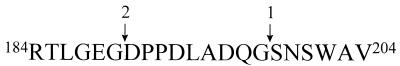Abstract
The glutaryl-7-aminocephalosporanic acid (GL-7-ACA) acylase of Pseudomonas sp. strain GK16 is an (αβ)2 heterotetramer of two nonidentical subunits. These subunits are derived from nascent polypeptides that are cleaved proteolytically between Gly198 and Ser199 after the nascent polypeptides have been translocated into the periplasm. The activation mechanism of the GL-7-ACA acylase has been analyzed by both in vivo and in vitro expression studies, site-directed mutagenesis, in vitro renaturation of inactive enzyme precursors, and enzyme reconstitution. An active enzyme complex was found in the cytoplasm when its translocation into the periplasm was suppressed. In addition, the in vitro-expressed GL-7-ACA acylase was processed into α and β subunits, and the inactive enzyme aggregate of the precursor was also processed and became active during the renaturation step. Mutation of Ser199 to Cys199 and enzyme reconstitution allowed us to identify the secondary processing site that resides in the α subunit and to show that Ser199 of the β subunit is essential for these two sequential processing steps. Mass spectrometry clearly indicated that the secondary processing occurs at Gly189-Asp190. All of the data suggest that the enzyme is activated through a two-step autocatalytic process upon folding: the first step is an intramolecular cleavage of the precursor between Gly198 and Ser199 for generation of the α subunit, containing the spacer peptide, and the β subunit; the second is an intermolecular event, which is catalyzed by the N-terminal Ser (Ser199) of the β subunit and results in a further cleavage and the removal of the spacer peptide (Asp190 to Gly198).
The glutaryl-7-aminocephalosporanic acid (GL-7-ACA) acylase (EC 3.5.1.11) of Pseudomonas sp. strain GK16 deacylates GL-7-ACA to 7-aminocephalosporanic acid (7-ACA), which is a starting material for the synthesis of semisynthetic cephem antibiotics (9, 10). The nascent polypeptide of the enzyme is synthesized as a 74-kDa polypeptide containing sequences coding for a signal peptide and 16-kDa α and 54-kDa β subunits, and the removal of the signal peptide gives rise to an inactive 70-kDa precursor, which has been suggested to be processed at a single site between Gly198 and Ser199 into 16-kDa α and 54-kDa β subunits in the periplasm (17). The active GL-7-ACA acylase is an (αβ)2 heterotetramer complex (10, 17).
The genes coding for this enzyme and other cephalosporin acylases from several Pseudomonas species have been cloned into Escherichia coli, and the enzymes were analyzed biochemically (1, 13, 14, 17, 18). The enzymes were efficiently processed and were found to be fully active even in a foreign host, E. coli. The enzymes are either a heterotetramer or a heterodimer of α and β subunits which are formed from each single-chain precursor protein. Noteworthy is the high conservation of the Ser residues at the N termini of the β subunits in both cephalosporin and penicillin acylases (1, 2, 6, 11, 14, 17, 18, 20, 21, 27). Recently, it was revealed that the N-terminal Ser residue of the β subunit of penicillin G acylase from E. coli could act as a nucleophile in both catalysis and processing and that the analogous residue in a cephalosporin C acylase from Pseudomonas sp. strain N176 might also be active in catalysis (4, 7, 11, 12). Other enzymes, so called Ntn (N-terminal nucleophile) hydrolases, have also been reported to be autocatalytically activated by the nucleophilic attack of the N-terminal amino acid residues of the β-subunits (4, 8, 15, 28, 30, 31).
To investigate the activation mechanism of the GL-7-ACA acylase precursor, we expressed the enzyme in a cell-free transcription-translation system. We also investigated the importance of a conserved Ser residue in the enzyme by site-directed mutagenesis and investigated the activation mechanism of the enzyme by the refolding of inactive precursors in vitro and enzyme reconstitution. Here, we report that the enzyme precursor autocatalytically converts to the α and β subunits and that a conserved Ser residue in the β subunit is necessary for both enzyme activity and processing. Furthermore, another processing site is found to exist in the α subunit, and the removal of a spacer peptide is required for the activation of the enzyme. Our data also imply that the enzyme is activated by a two-step autocatalytic mechanism, the first step of which is an intramolecular cleavage of the precursor at the primary processing site, generating an N-terminal Ser in the β subunit, and the second step of which is the intermolecular removal of a spacer peptide in the α subunit catalyzed by the N-terminal Ser of the β subunit.
MATERIALS AND METHODS
Recombinant DNA and mutagenesis.
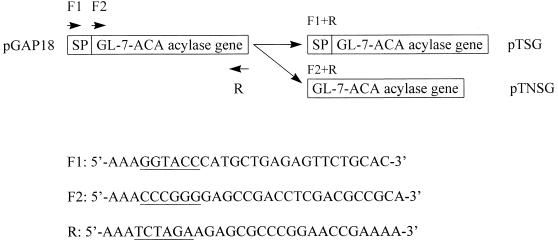
The plasmid pGAP18, harboring the cloned GL-7-ACA acylase gene from Pseudomonas sp. strain GK16 was described previously (14). The expression vector pTrxFus, containing the promoter λPL, and E. coli GI724 (Invitrogen) were used to construct two recombinant expression plasmids, pTSG and pTNSG (Fig. 1), which carry fusions of thioredoxin to the nascent and mature enzymes, respectively. The sequences and annealing sites of three primers used for PCR are shown in Fig. 1, and plasmid pGAP18 was used as a template. The PCR products encoding the nascent and mature GL-7-ACA acylases were cloned into the KpnI/SmaI and XbaI sites of the pTrxFus vector. For coupled in vitro transcription-translation reactions, each thioredoxin–GL-7-ACA acylase (TGA) fusion from the pTSG and pTNSG plasmids was subcloned into the NdeI and HindIII sites of the pET23a (Novagen) vector, containing the T7 RNA polymerase promoter. Site-directed mutations of Ser199 at the primary cleavage site of the enzyme precursor and a His6 tag at the C terminus of each fusion protein for its easy purification were introduced by using the megaprimer method (26) with a reverse primer containing an extra His6-coding sequence. As primers for site-directed mutagenesis and His6 tagging, the following oligonucleotides were used (changes are underlined): forward primer, 5′-GAGATATACATATGGCTAGCATGA-3′; mutagenic primers, 5′-CCCGGCGCCACCGCCCAGGAGTTGGCTCCTTGATC-3′ (for Ser199Ala mutation) and 5′-CCCGGCGCCACCGCCCAGGAGTTGCATCCTTGATC-3′ (for Ser199Cys mutation); and a reverse primer containing an extra His6-coding sequence, 5′-AAAAAGCTTTCAGTGATGGTGATGGTGATGTGGCTTGAAGTTGAAGGGCG-3′. For the wild-type fusion protein with a His6 tag, only the forward and reverse primers were used. Each amplified product was cloned into the NdeI and HindIII sites of the pET23a vector. All of the introduced mutations were confirmed by DNA sequence analyses (25). For the unfused enzyme with only the His6 tag, the following primers were used: forward primer, 5′-AAACCCGGGGAGCCGACCTCGACGCCGCA-3′, and reverse primer, 5′-AAAAAGCTTTCAGTGATGGTGATGGTGATGTGGCTTGAAGTTGAAGGGCG-3′. The PCR products coding for mature wild-type and Ser199Cys mutant enzymes with His6 tags were digested with SmaI and HindIII, cloned into a pET23d vector (Novagen) which was restricted with NcoI, filled in with Klenow enzyme, and subsequently digested with HindIII. All forward and reverse primers used in PCR had restriction enzyme sites at each 5′ end, and the gene manipulation applicable to E. coli was carried out as described by Sambrook et al. (24). All plasmids used for this study are described in Fig. 2.
FIG. 1.
Strategy for the construction of plasmids pTSG and pTNSG. Plasmid pGAP18 was used as a template in PCR. SP, DNA fragment coding for the signal peptide of GL-7-ACA acylase; F1 and F2, forward primers; R, reverse primer. The underlined sequences are restriction enzyme sites (KpnI, SmaI, and XbaI, respectively) attached to the annealing site of each primer.
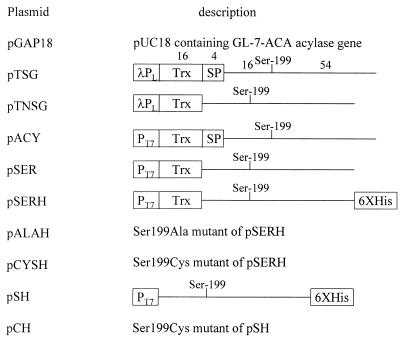
FIG. 2.
Recombinant plasmids used in this study. Numbers on the bars and lines denote the mass of each peptide or subunit, and the line stands for mature GL-7-ACA acylase gene. λPL, λPL promoter; Trx, thioredoxin-coding sequence; SP, signal peptide-coding sequence; T7, T7 promoter; Ser-199, Ser199 of a nascent GL-7-ACA acylase polypeptide or the N-terminal Ser residue of wild-type β subunit; 6XHis, sequence encoding six histidine residues. The plasmid pGAP18 has been described previously (14).
Enzyme assay.
The GL-7-ACA acylase assay was based on colorimetric measurement of 7-ACA released from the substrate GL-7-ACA as described previously (14). One unit of the enzyme releases 1 μmol of 7-ACA per min at 37°C under the enzyme assay conditions used.
In vitro transcription and translation reactions.
The various 35S-labeled proteins were made by using a coupled transcription and translation system (STP2; Novagen). Briefly, 0.5 mg of DNA was added directly to the STP2 T7 transcription mixture, and the reaction was carried out for 20 min at 30°C. The STP2 T7 transcription reaction mixture was then mixed with [35S]methionine and STP2 translation mixture and incubated for 1 h at 30°C, followed by a cold chase reaction with unlabeled methionine for 30 min at 30°C. The 35S-labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography.
Expression and purification of recombinant proteins.
E. coli GI724 cells harboring pTSG and pTNSG plasmids encoding TGA fusions were grown in induction medium (6 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 2 g of Casamino Acids, and 0.095 g of MgCl2 per liter) to an optical density at 550 nm of about 0.5 at 30°C and induced with tryptophan at a final concentration of 100 μg/ml for 2 h at 37°C. E. coli BL21(DE3) cells harboring each plasmid encoding wild-type and mutant thioredoxin enzyme fusions or mature enzymes with the C-terminal His6 tag were grown in Luria-Bertani medium at 37°C to an optical density at 550 nm of about 0.5 and induced with 0.4 mM isopropyl-1-thio-β-d-galactopyranoside for 2 h at 37°C. The cells were harvested by low-speed centrifugation (3,000 × g, 10 min). The cell pellets were resuspended in 20 mM Tris (pH 8.0)–100 mM NaCl–10% glycerol–0.1% Triton X-100 and lysed by sonication. The lysates were divided into soluble and insoluble fractions by high-speed centrifugation (15,000 × g, 20 min), and each fraction was used for the analysis and purification of expressed proteins.
Purification of native or denatured His6 proteins was performed by using TALON metal affinity resin (Clontech) according to the supplier’s instructions. The His6 proteins were purified by a batch method, and sonication buffer with 6 M urea was used for the purification of denatured His6 proteins. Each His6 protein was eluted with buffer containing 100 mM EDTA.
In vitro renaturation of inactive precursors.
Renaturation of purified inactive precursor was performed by a 15-fold dilution with renaturation buffer (20 mM Tris-HCl [pH 8.0] and 100 mM NaCl) at 25°C. The refolding mixtures were subjected to SDS-PAGE, and in vitro processing was examined by staining with Coomassie blue R-250 or Western blot analysis.
Reconstitution of enzymes.
For purification of the thioredoxin–α subunit of GL-7-ACA acylase (TGα) and β subunit of GL-7-ACA acylase with a His6 tag (Hβ) subunits from wild-type and Ser199Cys mutant fusion proteins, purified native fusion proteins were dialyzed against 20 mM Tris-HCl (pH 8.0)–100 mM NaCl–10% glycerol–0.1% Triton X-100 for the removal of EDTA and subsequently concentrated by using a Microcon 10 (Amicon). After the subunits of the fusion proteins were dissociated with 6 M urea, they were purified with TALON resin on the basis of the fact that only the Hβ subunit can bind to the resin. The protein concentration was determined by the Bradford method (3) with bovine serum albumin as a standard. For enzyme reconstitution, each denatured and purified subunit was mixed at a molar ratio of 1:1 in cis (within the same precursor) or in trans (between the wild type and the Ser199Cys mutant), and diluted 15-fold with renaturation buffer at 25°C for 12 h so that the enzymes could be reconstituted.
Western blot analysis.
Western blotting was carried out by the procedure described by Towbin et al. (33). The proteins on SDS-PAGE (10% acrylamide) were electroblotted to nitrocellulose paper and detected by anti-Thio and anti-His6 mouse monoclonal antibodies (commercially available from Invitrogen and Clontech, respectively). Sheep anti-mouse immunoglobulin G conjugated with horseradish peroxidase and ECL detection reagents (Amersham) were used as the secondary antibody and the detection solutions, respectively.
N-terminal amino acid sequencing and determination of C-terminal amino acid residue.
N-terminal amino acid sequence determination was performed on an Applied Biosystems 491 protein sequencer fitted with a high-pressure liquid chromatography on-line system. Purified native and in vitro-refolded fusion proteins of the wild type and the Ser199Cys mutant were subjected to electrophoresis on a 10% polyacrylamide gel and electroblotted to a polyvinylidine difluoride membrane according to the procedure of Matsudaira (19). The membrane was stained with Coomassie blue R-250, and the protein bands of the Hβ subunit were excised and subjected to sequential Edman degradation. The C-terminal amino acid of the wild-type α subunit was determined as follows: (i) purification of α subunits from wild-type and Ser199Cys mutant nonfusion proteins, (ii) measurement of the mass of each α subunit by MALDI (matrix-assisted laser desorption-ionization) mass spectrometry with a Kratos Kompact MALDI II instrument (29), and (iii) comparison of the measured masses with molecular masses calculated from the deduced amino acid sequence of the α subunit.
RESULTS
Construction and expression of TGA fusions in the cytoplasm.
The GL-7-ACA acylase is a periplasmic enzyme in Pseudomonas sp. strain GK16 and has also been found to be correctly processed and active in the periplasm when the cloned gene is expressed in E. coli (14, 17). These data indicate that after a nascent polypeptide is translocated into the periplasm, processing and heterotetramer formation of the active enzyme take place in this compartment and that a common activation mechanism or machinery must operate in both Pseudomonas sp. strain GK16 and E. coli. To examine this process further, we constructed two recombinant TGA fusion plasmids, pTSG and pTNSG; the former contains the gene encoding the whole enzyme, including its 29-amino-acid signal peptide sequence, and the latter has only a DNA fragment encoding the mature enzyme. The thioredoxin moiety appears not only to confer high solubility on heterologous proteins but also to enable fusion proteins to localize to the adhesion zones or Bayer’s patches (16).
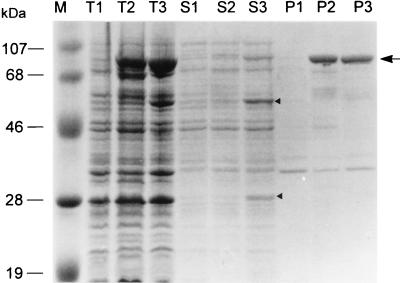
Only the soluble fraction of a cell extract from cells with pTNSG contained a detectable level of two polypeptides generated from the cleavage of the fusion precursor (Fig. 3); one is the 32-kDa fusion protein between thioredoxin and the α subunit, termed TGα, and the other is the normal β subunit with a mass of 54 kDa (lane S3). No processing could be detected in the inactive protein aggregates from cells with pTNSG and pTSG (Fig. 3, lanes P2 and P3). The inefficiency of the processing of soluble fusion proteins generated from pTSG (lane S2) may be due to the existence of a signal peptide sequence between thioredoxin and mature GL-7-ACA acylase, and the signal peptide was suggested to play a role in maintaining a translocating protein in an unfolded state (22, 23). The soluble fraction from cells with pTNSG had an approximately 3.5-fold-higher enzyme activity than the same fraction from cells with pTSG (Table 1), suggesting that enzyme activity is related to the degree of proteolytic processing of the enzyme precursor and that its proteolytic processing may occur upon protein folding in the absence of signal peptide. The enzymatically active fusion protein expressed from pTNSG was localized within the osmotically sensitive cellular compartment, the adhesion zone, and not in the periplasm (data not shown). This indicates that although the fusion protein expressed from pTNSG was confined to the cytoplasm, it could be activated in this cellular compartment and that the proteolytic cleavage of the fusion precursor might occur by an intrinsic property of the precursor, such as autocatalytic processing.
FIG. 3.
SDS-PAGE analyses of the expression and processing patterns of TGA fusion proteins. Lane M, molecular mass markers; lanes T1 to T3, total cell extracts; lanes S1 to S3, soluble fractions; lanes P1 to P3, insoluble fractions from pTrxFus (vector)-, pTSG-, and pTNSG-harboring E. coli GI724, respectively. The upper and lower arrowheads in lane S3 denote the processed 54-kDa β subunit and 32-kDa TGα subunit, respectively. The arrow indicates each unprocessed 86-kDa protein aggregate.
TABLE 1.
GL-7-ACA acylase activities of TGA fusion proteins expressed from pTSG- and pTNSG-harboring E. coli GI724 cells
| Plasmid | Activity (U/ml of culture broth) in:
|
|
|---|---|---|
| Soluble fraction | Insoluble fraction | |
| pTrxFus (vector) | NDa | ND |
| pTSG | 0.29 | 0.24 |
| pTNSG | 0.95 | 0.29 |
ND, not detectable.
Processing of in vitro-transcribed and -translated TGA fusion proteins.
We could not exclude the possibility that Pseudomonas sp. strain GK16 and E. coli possess a common protease responsible for the proteolytic cleavage of GL-7-ACA acylase. To determine whether the activation of the TGA fusion protein is an autoproteolytic process, we synthesized the two TGA fusion proteins described above by using in vitro- coupled transcription and translation reactions. As shown in lanes 2 and 3 of Fig. 4, even in vitro-expressed fusion precursors were normally cleaved into smaller fragments. As found in vivo, a TGA fusion precursor containing a signal peptide was also cleaved at a lower rate than the precursor without the signal peptide in vitro. This result strongly suggests that the enzyme is cleaved through an autocatalytic process, since the in vitro expression system most likely does not contain any protease able to catalyze this cleavage.
FIG. 4.
Processing patterns of in vitro-expressed TGA fusion proteins in coupled in vitro transcription and translation reactions. Lane 1, in vitro-expressed β-galactosidase as a positive control; lanes 2 and 3, in vitro-expressed TGA fusion proteins from plasmids pACY and pSER, respectively. The upper and lower arrowheads in lanes 2 and 3 denote the processed β subunits and the TGα subunits, respectively, while the arrow denotes each unprocessed TGA fusion precursor. The mass of the TGα subunit in lane 2 is slightly higher than that of the TGα subunit in lane 3 because of the presence of the signal peptide in the fusion.
Effect of substitutions for Ser199 on the catalytic and processing properties of a TGA fusion protein.
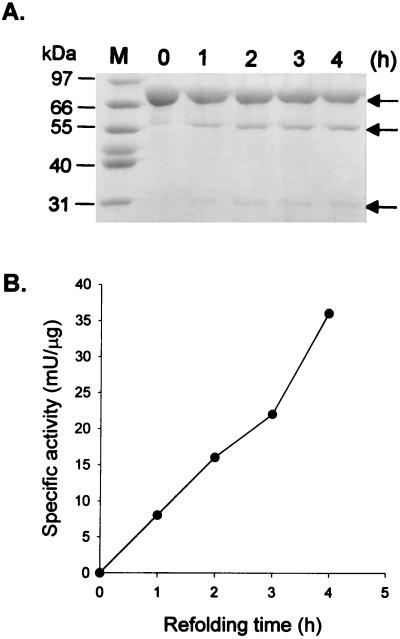
We chose a TGA fusion protein without a signal peptide sequence for the further study of the activation mechanism of GL-7-ACA acylase, since it was processed at an appropriate rate. In addition, we tagged six His residues at the C terminus of the TGA fusion protein for its easy purification with TALON affinity resin. The TGA fusion protein containing the His6 tag is termed TGAH. To confirm that the TGA fusion protein is maturated by an autocatalytic process, we expressed the TGAH fusion protein in E. coli and isolated the inactive precursor as a denatured protein. When the purified TGAH precursor was refolded, enzyme activity was regained and the precursor was cleaved into smaller fragments, one of which is the 32-kDa TGα and the other of which is the 54-kDa Hβ (Fig. 5). The enzyme activity was regained in proportion to the degree of processing of the TGAH precursor. The result demonstrates that a specific processing intermediate does not exist (Fig. 5A) and that the processing occurs through autocatalysis upon folding and is dependent on the proper folding of the precursor.
FIG. 5.
Time course of in vitro processing of inactive wild-type TGAH fusion precursor expressed from plasmid pSERH (A) and enzyme activity as a function of refolding time (B). (A) Purified inactive wild-type TGAH fusion precursor was renatured at each indicated time and immediately mixed and boiled for 10 min with sample application buffer for SDS-PAGE. The conversion of the TGAH fusion precursor to TGα and Hβ subunits was monitored by SDS-PAGE. Lane M, molecular mass markers. The arrows, from the top to the bottom, indicate unprocessed fusion precursor, Hβ subunit, and TGα subunit, respectively. (B) The enzyme activity of each refolding mixture at various times was assayed as described in Materials and Methods.
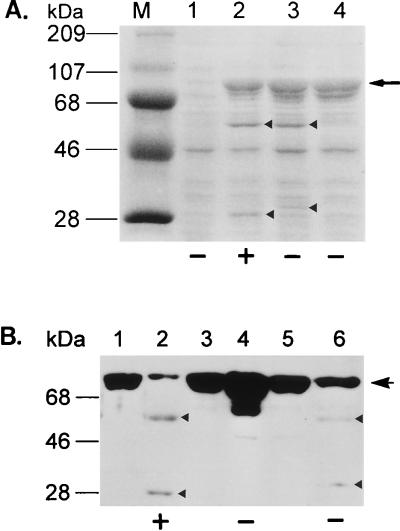
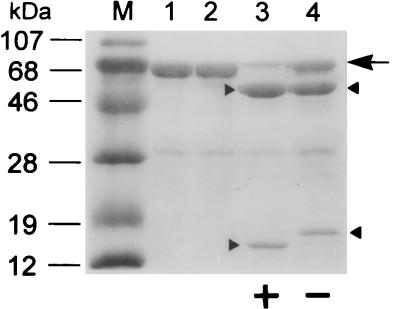
Alignment of the amino acid sequences of the GL-7-ACA acylase with those of other known cephalosporin and penicillin acylases identified a conserved Ser at the processing site of each enzyme (1, 2, 6, 11, 14, 17, 18, 20, 21, 27); after processing, this Ser is at the N terminus of the β subunit. To study the role of Ser199 in catalysis and in the maturation process, we mutagenized this residue to either Ala or Cys. Strains carrying the mutations exhibited no enzyme activity, suggesting a functional role of Ser199 similar to that of Ser290 of penicillin G acylase. SDS-PAGE analysis with TGAH fusion proteins expressed in vivo showed that the Ser199Cys mutant fusion precursor was cleaved proteolytically, resulting in a normal Hβ subunit and a TGα polypeptide slightly larger than that of the wild type (Fig. 6A, lane 3), whereas the replacement of Ser199 with Ala blocked the cleavage of the TGAH fusion precursor (Fig. 6A, lane 4). Processing and enzyme activity of renatured wild-type and mutant fusion precursors were similar to those of in vivo-expressed proteins (Fig. 6B). N-terminal sequence analysis of the wild-type and Ser199Cys Hβ subunits generated from in vivo purification and in vitro processing showed that the processing was at the right place, i.e., at the N-terminal side of residue 199 (data not shown). To exclude any artificial effect of thioredoxin on the enzyme activity and processing, we made both wild-type and Ser199Cys variants to which only the His6 tag was attached at the C terminus, purified the precursors, and examined protein refolding. In vitro processing of both the wild-type and Ser199Cys precursors of these latter fusions was the same as that of the TGAH fusions, showing that the presence of thioredoxin did not affect autoproteolysis and enzyme activity (Fig. 7). In vivo processing and enzyme activities were also the same with the two types of fusions (data not shown). To determine whether the proteolytic activity of the enzyme could act in trans, purified active wild-type TGAH fusion protein was added to a refolding mixture of inactive wild-type precursor, and an aliquot of the protein sample was withdrawn at various times and analyzed for enhanced processing rate of the wild-type precursor. The result showed that the proteolytic cleavage of the enzyme precursor must occur only in cis in a folding-dependent manner, since the processing rate of the inactive wild-type precursor was not enhanced by active enzyme (data not shown). These results also indicate that a hydroxyl or sulfhydryl side chain at position 199 may be required for processing of the precursor. The fact that only the wild-type fusion protein, and not the Ser199Cys mutant, retained enzyme activity suggests that a hydroxyl residue at the N terminus of the β subunit is required for enzyme activity. Thus, the hydroxyl residue of Ser199 could be involved in processing as well as in the enzyme activity.
FIG. 6.
In vivo (A) and in vitro (B) processing patterns of wild-type and mutant TGAH fusion precursors analyzed by SDS-PAGE and immunoblotting, respectively. (A) Lane M, molecular mass markers; lane 1, total cell lysate from cells with vector pET23a as a negative control; lanes 2 to 4, total cell lysates from cells with plasmids pSERH, pCYSH, and pALAH, respectively. From top to bottom, the arrowheads in lanes 2 and 3 denote the Hβ and TGα subunits, respectively. Note that the TGα subunit generated from plasmid pCYSH is slightly larger than that of the wild type (lane 3). The arrow indicates each expressed protein aggregate. + and −, presence and absence of enzyme activity, respectively. (B) Immunoblotting of purified inactive precursors and renatured proteins with monoclonal antibodies against thioredoxin and His6 tag, respectively. Lanes 1, 3, and 5, before renaturation; lanes 2, 4, and 6, after renaturation of the precursors expressed from plasmids pSERH, pALAH, and pCYSH, respectively. The upper arrow denotes each purified inactive fusion precursor, and the upper and lower arrowheads in lanes 3 and 6 indicate Hβ and TGα subunits, respectively. + and −, acquisition or total lack of enzyme activity, respectively.
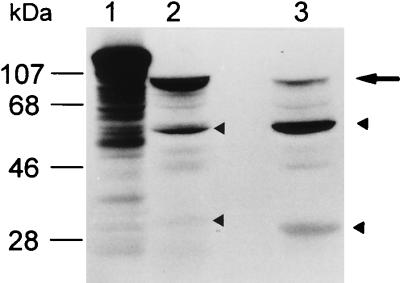
FIG. 7.
In vitro processing pattern of unfused inactive wild-type and Ser199Cys mutant GL-7-ACA acylase precursors. Unfused precursors expressed from pSH and pCH were renatured and then subjected to SDS-PAGE. Lane M, molecular mass markers; lanes 1 and 2, before renaturation; lanes 3 and 4, after renaturation of the unfused precursors expressed from plasmids pSH and pCH, respectively. The large arrow denotes each purified inactive precursor, and the upper and lower arrowheads indicate the Hβ subunit and the α subunit containing extra amino acids, Met and Gly, at its N terminus, respectively. + and −, acquisition or total lack of enzyme activity, respectively.
Enzyme reconstitution and identification of secondary processing site.
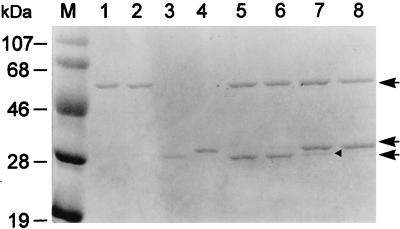
As described in the previous section, replacement of Ser199 with Cys resulted in a TGα polypeptide slightly larger than that of the wild type generated by proteolytic cleavage. Since N-terminal sequence analysis revealed that cleavage occurred correctly at the N-terminal side of residue 199 of wild-type or Ser199Cys mutant GL-7-ACA acylase, the larger TGα polypeptide from the Ser199Cys mutant may result from a lack of cleavage at another processing site existing in the α subunit of the enzyme. The lack of enzyme activity of the Ser199Cys mutant fusion protein might then be due to either the incomplete processing of the TGα subunit or the Cys residue at the N terminus of its Hβ subunit. To test these possibilities, we purified the TGα and Hβ subunits from wild-type and Ser199Cys mutant fusion proteins in the denatured condition and reconstituted the enzyme by refolding. The enzyme was active when it was reconstituted with the combination of TGα and Hβ subunits derived from the wild-type fusion protein but not when it was reconstituted with the normal TGα subunit from the wild type and the Hβ subunit from Ser199Cys mutant, indicating that the hydroxyl residue of Ser199 was necessary for the enzyme activity (Fig. 8, lanes 5 and 6; Table 2). In the reconstitution of the larger TGα subunit with either wild-type or Ser199Cys mutant Hβ subunit, only the former reconstitution restored enzyme activity, with a partial conversion of the larger TGα subunit to the normal wild-type one (Fig. 8, lanes 7 and 8; Table 2). This demonstrated that the removal of a spacer peptide was required for the enzyme activity and that the hydroxyl residue of Ser199 played an essential role in this secondary processing as well as in the primary one.
FIG. 8.
Protein profile of each reconstituted TGAH fusion protein. Lane M, molecular mass markers; lanes 1 and 3, purified Hβ and TGα subunits from plasmid pSERH; lanes 2 and 4, purified Hβ and TGα subunits from plasmid pCYSH; lane 5, reconstitution of Hβ and TGα subunits from plasmid pSERH; lane 6, reconstitution of Hβ subunit from plasmid pCYSH with TGα subunit from plasmid pSERH; lane 7, reconstitution of Hβ subunit from plasmid pSERH with TGα subunit from plasmid pCYSH; lane 8, reconstitution of Hβ and TGα subunits from plasmid pCYSH. From top to bottom, the arrows indicate the Hβ subunits from either pSERH or pCYSH, the TGα subunit from plasmid pCYSH, and the TGα subunit from plasmid pSERH, respectively. The arrowhead indicates a further processed TGα subunit from plasmid pCYSH.
TABLE 2.
GL-7-ACA acylase activities of reconstituted enzymes
| Component(s)a | Sp act (U/μg) | % Sp actb |
|---|---|---|
| WT TGα + WT Hβ | 0.012 | 19 |
| WT TGα + mutant Hβ | 0 | 0 |
| Mutant TGα + WT Hβ | 0.011 | 17.5 |
| Mutant TGα + mutant Hβ | 0 | 0 |
| Active TGAH protein | 0.063 | 100 |
WT, wild type. Mutant refers to subunits of Ser199Cys-mutated TGAH protein. Active TGAH protein was used for estimating the efficiency of enzyme reconstitution.
The percent specific activity of each reconstituted enzyme was calculated by comparison with the specific activity of active TGAH protein.
The secondary processing site of the GL-7-ACA acylase precursor was identified by analysis of the normal and larger α subunits purified from nonfusion wild-type and Ser199Cys mutant enzyme by using MALDI mass spectrometry. The secondary processing site was located between Gly189 and Asp190 in the enzyme precursor (Fig. 9).
FIG. 9.
Amino acid sequence showing the primary (arrow 1) and secondary (arrow 2) processing sites in the GL-7-ACA acylase precursor.
Inhibition studies of GL-7-ACA acylase.
The studies of Ser199Cys substitution and reconstitution clearly indicated the essential role of Ser199 of the β subunit. To examine whether the enzyme activity and the autoproteolytic activation of GL-7-ACA acylase are inhibited by serine protease inhibitors, GL-7-ACA acylase, its precursor, penicillin G acylase, and other serine protease-type enzymes were treated with the inhibitors (Table 3). In all cases the activity of trypsin was completely inhibited. Interestingly, the enzyme activity of penicillin G acylase was completely inhibited with phenylmethylsulfonyl fluoride (PMSF) (30), whereas GL-7-ACA acylase activity was not affected. Diisopropylphosphofluoridate (DFP), which is a potent inhibitor of serine protease, inhibited neither GL-7-ACA acylase nor penicillin G acylase. The autoproteolytic activation of GL-7-ACA acylase was also not affected by the inhibitors.
TABLE 3.
Inhibition of GL-7-ACA acylasea
| Inhibitorb | Activity or processingc
|
|||
|---|---|---|---|---|
| GL-7-ACA acylase | Penicillin G acylase | Subtilisin | Trypsin | |
| DFP | + | + | − | − |
| PMSF | + | − | − | − |
| APMSF | + | + | + | − |
| 3,4-DCI | + | + | + | − |
| Leupeptin | + | + | + | − |
Studies of the inhibition of autocatalytic processing and activity of GL-7-ACA acylase were performed with the precursor and active forms of wild-type TGAH protein, respectively. Penicillin G acylase, subtilisin, and trypsin from E. coli ATCC 11105, Bacillus subtilis, and bovine pancreas, respectively, were used as controls for inhibition of enzyme activity. Except for 3,4-DCI (1 mM), all compounds were tested at a final concentration of 5 mM. Prior to incubation with appropriate substrates or refolding in the presence of 1 or 5 mM inhibitor, the enzymes or precursor were preincubated with each inhibitor.
APMSF, 4-amidinophenylmethylsulfonyl fluoride; 3,4-DCI, 3,4-dichloroisocoumarin.
−, complete absence of enzyme activity; +, presence of enzyme activity and, for GL-7-ACA acylase, normal processing of the TGAH precursor.
DISCUSSION
The correct processing and formation of GL-7-ACA acylase in the cytoplasm suggested to us that the activation of the enzyme might be an intrinsic property of the protein. This was further suggested by proper cleavage of the precursor translated in the rabbit reticulocyte lysate system and the complete processing and gain of enzymatic activity upon renaturation of the purified inactive precursor.
It has been suggested that the GL-7-ACA acylase precursor of Pseudomonas sp. strain GK16 has a single cleavage site located between Gly198 and Ser199 for the generation of α and β subunits (17). In this study, we have identified a secondary processing site upstream of the primary processing site in the precursor. The secondary processing site is located between Gly189 and Asp190, and the removal of a nine-amino-acid spacer peptide is necessary for the activation of the enzyme. Interestingly, the Gly-Asp sequence is also present with the spacing of eight amino acids from the Ser residue at the primary processing site, Gly-Ser, in the cephalosporin C acylase precursor of Pseudomonas sp. strain SE83 (18), although it is unknown whether this cephalosporin C acylase also requires secondary processing for the activation of the enzyme. It will be of interest to see if this cephalosporin C acylase precursor is processed by autoproteolysis and undergoes the secondary processing between Gly and Asp for the activation of the enzyme. It has been reported that the absence of a signal peptide in the penicillin G acylase precursor blocked the secondary processing of the α subunit, removing a spacer peptide, and decreased the primary processing efficiency of the β subunit (5). In contrast, in this study, the absence of a signal peptide in the enzyme precursor did not prevent the precursor from removing a spacer peptide by complete autoproteolysis both in vivo and in vitro and suggested that the processing was folding related as well (Fig. 7).
In this study we have identified the hydroxyl group of Ser199 of GL-7-ACA acylase as being involved in autoproteolysis and enzyme activity. The replacement of a highly conserved amino acid residue, Ser199, with Cys resulted in an incomplete processing in which the proteolytic cleavage took place only at the primary processing site both in vivo and in vitro and consequently resulted in a total loss of GL-7-ACA acylase activity. The in vitro primary processing rate of the Ser199Cys mutant precursor was lower than that of the wild type (Fig. 7). However, it should be noted that the primary processing is very specific; the NH2-terminal amino acid sequence of the β subunit of the Ser199Cys mutant is identical to that of the wild type except for the first amino acid (Cys for Ser). The fact that the hydroxyl or sulfhydryl group of residue 199 is required for the primary processing led us to think that the specific cleavage is mediated by the nucleophilic attack of the side chain of residue 199, just as the hydroxyl group of serine protease cleaves the substrate by its nucleophilic attack. However, enzyme reconstitution experiments showed that a Cys at residue 199 does not catalyze the secondary processing step. If the hydroxyl or sulfhydryl groups of residue 199 in wild-type and Ser199Cys mutant enzyme precursors act only as a nucleophile in the processing reactions, the decreased primary processing rate and lack of secondary processing and enzyme activity of the Ser199Cys mutant must presumably result from a defect such as structural hindrance due to the physical properties of the sulfhydryl group, since the sulfhydryl group of cysteine has a higher nucleophilicity than the hydroxyl group of serine.
In contrast to the lack of secondary processing due to the sulfhydryl group of residue 199 in the enzyme precursor, replacement of the corresponding Ser residue in the penicillin G acylase precursor with Cys allowed both secondary and primary processing of the precursor but resulted in the loss of enzyme activity (5). Since the variant in which Ser199 was replaced by Ala gave neither processing nor enzyme activity, our results unambiguously demonstrate that Ser199 at the primary processing site plays an active role in the complete processing of autoproteolysis as well as in enzyme activity. The results also suggest that the GL-7-ACA acylase precursor is processed through two sequential steps of autocatalysis, the first being an intramolecular cleavage of the precursor at the primary processing site for the generation of the N-terminal Ser199 residue of the β subunit and the second being an intermolecular cleavage for the removal of the spacer peptide in the α subunit catalyzed by the N-terminal Ser-199 of the β subunit.
Penicillin G acylase, proteasome β subunit, glutamine amidotransferase, and aspartylglucosaminidase have recently been suggested to form a novel class of Ntn (N-terminal nucleophile) hydrolases. All of these hydrolases have a common structural motif and an N-terminal catalytic amino acid: Ser in penicillin G acylase, Thr in proteasome β subunit and aspartylglucosaminidase, and Cys in glutamine amidotransferase. It has been suggested that the α amino group of the N-terminal catalytic amino acid could function as a base that increases the nucleophilicity of the hydroxyl or sulfhydryl group. In addition, autocatalytic processing from an inactive precursor polypeptide seems to be common to these hydrolases. In the case of penicillin G acylase, 1 mol of PMSF/mol of the enzyme completely and rapidly inactivated the enzyme (30), but it has not been reported that the processing of the enzyme precursor is inhibited under the same conditions. Unexpectedly, both PMSF and DFP did not inhibit the enzyme activity and autoproteolytic maturation of GL-7-ACA acylase, although penicillin G acylase, which belongs to the Ntn hydrolase superfamily, used as a control was rapidly and completely inactivated with PMSF. Our current data do not clearly indicate the exact role of the N-terminal Ser residue of the β subunit, since the Ser residue is essential in the substitution studies but it is not a catalytic center as judged by the lack of inhibition by several serine protease inhibitors, such as DFP, PMSF, and 4-amidinophenylmethylsulfonyl fluoride. At this point, it is not clear whether the enzyme is a member of the Ntn hydrolase superfamily or not. We believe, however, that the enzyme may be a member of the Ntn hydrolase superfamily, since it is highly homologous to penicillin G acylase and the N-terminal Ser199 of the β subunit is essential for the autoproteolytic activation and catalytic activity of the enzyme. Crystallization of the enzyme is under way. We expect that the crystal structure of the enzyme will help us solve this problem.
This study is the first characterization of the autoproteolytic activation of GL-7-ACA acylase, a cephalosporin acylase. In case of aspartylglucosaminidase, an (αβ)2 heterotetramer, dimerization of two inactive precursor molecules has been suggested to be the triggering event resulting in the autoproteolytic activation (32). It remains to be determined what the structural motif of GL-7-ACA acylase is and whether the dimerization of two inactive enzyme precursors precedes the autocatalytic processing of the precursor. It is obvious, however, that the processing of the enzyme occurs within a fold in cis, since neither the active enzyme nor β subunit which was added during in vitro renaturation of the enzyme precursor increased the processing rate in trans. The detailed autoproteolytic mechanism for the activation of GL-7-ACA acylase also remains to be determined.
ACKNOWLEDGMENTS
This research was supported by the Highly Advanced Project of the Ministry of Science and Technology, Republic of Korea.
We are very grateful to Roy H. Doi, University of California, Davis, for his critical comments and discussion of the manuscript.
REFERENCES
- 1.Aramori I, Fukagawa M, Tsumura M, Iwami M, Isogai T, Ono H, Ishitani Y, Kojo H, Kohsaka M, Ueda Y, Imanaka H. Cloning and nucleotide sequencing of new glutaryl 7-ACA and cephalosporin C acylase genes from Pseudomonas strain. J Ferment Bioeng. 1991;72:232–243. [Google Scholar]
- 2.Barbero J L, Buesa J M, de Buitrago G G, Méndez E, Pérez-Aranda A, García J L. Complete nucleotide sequence of the penicillin acylase gene from Kluyvera citrophilia. Gene. 1986;49:69–80. doi: 10.1016/0378-1119(86)90386-0. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Brannigan J A, Dodson G, Duggleby H J, Moody P C E, Smith J L, Tomchick D R, Murzin A G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 5.Choi K S, Kim J A, Kang H S. Effects of site-directed mutations on processing and activities of penicillin G acylase from Escherichia coli ATCC 11105. J Bacteriol. 1992;174:6270–6276. doi: 10.1128/jb.174.19.6270-6276.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daumy G O, Danley D, McColl A S. Role of protein subunits in Proteus rettgeri penicillin G acylase. J Bacteriol. 1985;163:1279–1281. doi: 10.1128/jb.163.3.1279-1281.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duggleby H J, Tolley S P, Hill C P, Dodson E J, Dodson G, Moody P C E. Penicillin acylase has a single-amino-acid catalytic centre. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 8.Guan C, Cui T, Rao V, Liao W, Benner J, Lin C, Comb D. Activation of glycosylasparaginase. J Biol Chem. 1996;271:1732–1737. doi: 10.1074/jbc.271.3.1732. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa S, Murai Y, Yamamoto S, Shibuya Y, Fujii T, Komatsu K, Kodaira R. The isolation and properties of Pseudomonas mutants with enhanced productivity of 7β-(4-carboxybutanamido)cephalosporanic acid acylase. Agric Biol Chem. 1981;45:2225–2229. [Google Scholar]
- 10.Ichikawa S, Shibuya Y, Matsumoto K, Fujii T, Komatsu K, Kodaira R. Purification and properties of 7β-(4-carboxybutanamido)cephalosporanic acid acylase produced by mutants derived from Pseudomonas. Agric Biol Chem. 1981;45:2231–2236. [Google Scholar]
- 11.Ishii Y, Saito Y, Sasaki H, Uchiyama F, Hayashi M, Nakamura S, Niwa M. Affinity labeling of cephalosporin C acylase from Pseudomonas sp. N176 with substrate analogue, 7β-(6-bromohexanoylamido)cephalosporanic acid. J Ferment Bioeng. 1994;77:598–603. [Google Scholar]
- 12.Ishii Y, Saito Y, Fujimura T, Sasaki H, Noguchi Y, Yamada H, Niwa M, Shimomura K. High-level production, chemical modification and site-directed mutagenesis of a cephalosporin C acylase from Pseudomonas strain N176. Eur J Biochem. 1995;230:773–778. [PubMed] [Google Scholar]
- 13.Ishii Y, Saito Y, Fujimura T, Isogai T, Kojo H, Yamashita M, Niwa M, Kohsaka M. A novel 7-β-(4-carboxybutanamido)-cephalosporanic acid acylase isolated from Pseudomonas strain C427 and its high-level production in Escherichia coli. J Ferment Bioeng. 1994;77:591–597. [Google Scholar]
- 14.Lee Y S, Yang H C, Park S S. Cloning and characterization of GL-7-ACA acylase gene from Pseudomonas sp. GK16. J Microbiol Biotechnol. 1996;6:375–380. [Google Scholar]
- 15.Löwe J, Stock D, Jap B, Zwickle P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 16.Lunn C A, Pigiet V P. Localization of thioredoxin from Escherichia coli in an osmotically sensitive compartment. J Biol Chem. 1982;257:11424–11430. [PubMed] [Google Scholar]
- 17.Matsuda A, Komatsu K. Molecular cloning and structure of the gene for 7β-(4-carboxybutanamido)cephalosporanic acid acylase from a Pseudomonas strain. J Bacteriol. 1985;163:1222–1228. doi: 10.1128/jb.163.3.1222-1228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda A, Matsuyama K, Yamamoto K, Ichikawa S, Komatsu K. Cloning and characterization of the genes for two distinct cephalosporin acylases from a Pseudomonas strain. J Bacteriol. 1987;169:5815–5820. doi: 10.1128/jb.169.12.5815-5820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 20.Oh S J, Kim Y C, Park Y W, Min S Y, Kim I S, Kang H S. Complete nucleotide sequence of the penicillin G acylase gene and the flanking regions, and its expression in Escherichia coli. Gene. 1987;56:87–97. doi: 10.1016/0378-1119(87)90161-2. [DOI] [PubMed] [Google Scholar]
- 21.Ohashi H, Katsuta Y, Hashizume T, Abe S, Kajiura H, Hattori H, Kamei T, Yano M. Molecular cloning of the penicillin G acylase gene from Arthrobacter viscosus. Appl Environ Microbiol. 1988;54:2603–2607. doi: 10.1128/aem.54.11.2603-2607.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randall L L, Hardy S J S. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 23.Randall L L, Hardy S J S. Unity in function in the absence of consensus in sequence: role of leader peptides in export. Science. 1989;243:1156–1159. doi: 10.1126/science.2646712. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 27.Schumacher G, Sizmann D, Haug H, Buckel P, Böck A. Penicillin acylase from E. coli: unique gene-protein relation. Nucleic Acids Res. 1986;14:5713–5727. doi: 10.1093/nar/14.14.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemüller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 29.Siuzdak G. The emergence of mass spectrometry in biochemical research. Proc Natl Acad Sci USA. 1994;91:11290–11297. doi: 10.1073/pnas.91.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slade A, Horrocks A J, Lindsay C D, Dunbar B, Virden R. Site-directed chemical conversion of serine to cysteine in penicillin acylase from Escherichia coli ATCC 11105. Eur J Biochem. 1991;197:75–80. doi: 10.1111/j.1432-1033.1991.tb15884.x. [DOI] [PubMed] [Google Scholar]
- 31.Smith J L, Zaluzec E J, Wery J P, Niu L, Switzer R L, Zalkin H, Satow Y. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science. 1994;264:1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- 32.Tikkanen R, Riikonen A, Oinonen C, Rouvinen J, Peltonen L. Functional analyses of active site residues of human lysosomal aspartylglucosaminidase: implications for catalytic mechanism and autocatalytic activation. EMBO J. 1996;15:2954–2960. [PMC free article] [PubMed] [Google Scholar]
- 33.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]