Abstract
R1Bm is a non-LTR retrotransposon found specifically within 28S rRNA genes of the silkworm. Different from other non-LTR retrotransposons encoding two open reading frames (ORFs), R1Bm structurally lacks a poly (A) tract at its 3′ end. To study how R1Bm initiates reverse transcription from the poly (A)-less template RNA, we established an in vivo retrotransposition system using recombinant baculovirus, and characterized retrotransposition activities of R1Bm. Target-primed reverse transcription (TPRT) of R1Bm occurred from the cleavage site generated by endonuclease (EN). The 147 bp of 3′-untranslated region (3′UTR) was essential for efficient retrotransposition of R1Bm. Even using the complete R1Bm element, however, reverse transcription started from various sites of the template RNA mostly with 5′-UG-3′ or 5′-UGU-3′ at their 3′ ends, which are presumably base-paired with 3′ end of the EN-digested 28S rDNA target sequence, 5′-AGTAGATAGGGACA-3′. When the downstream sequence of 28S rDNA target was added to the 3′ end of R1 unit, reverse transcription started exactly from the 3′ end of 3′UTR and retrotransposition efficiency increased. These results indicate that 3′-terminal structure of template RNA including read-through region interacts with its target rDNA sequences of R1Bm, which plays important roles in initial process of TPRT in vivo.
INTRODUCTION
Non-long terminal repeat (non-LTR) retrotransposons, also called long interspersed nuclear elements (LINEs) or poly (A) elements, are the most abundant family among mobile elements. LINEs have been identified in all major groups of eukaryotes, with the exception of the bdelloid rotifers (1). In humans, up to 21% of the genome is comprised of LINEs (2), and the involvement of LINEs in the gene evolution and genome reconstruction had been proposed (3–5).
LINEs can be classified into two subtypes based on their structures and modes of retrotransposition (6). The early-branched subtype, such as R2 element specifically integrated in 28S rDNA of arthropods, comprises a single open reading frame (ORF) that encodes reverse transcriptase (RT) in the middle and a restriction enzyme-like endonuclease (EN) near its C-terminal end (7,8). Biochemical analyses of R2Bm of the silkworm led to the current model for LINE retrotransposition. R2Bm protein first recognizes the 3′UTR sequence of R2 RNA, and makes a specific nick on the target DNA by its endonuclease (9,10). Then, the exposed 3′ hydroxyl is utilized to initiate reverse transcription of the RNA template (11). Thus, the processes of nicking, reverse transcription and integration are basically coupled, which are termed target-primed reverse transcription (TPRT). It is suggested that the rare transcript of R2 element is provided by co-transcription with the 28S rDNA target (12,13), and an additional 28S rRNA sequence downstream of the R2 sequence reduced the efficiency of the reaction, but increased the rate of accurate junctions seen in vivo (14).
The recently-branched subtype of LINEs has two ORFs: ORF1 and ORF2. The ORF2 encodes catalytic domains responsible for LINE retrotransposition; apurinic/apyrimidinic endonucleases (APE)-like endonuclease at the N-terminus (15) and reverse transcriptase at the central region. Retrotransposition of this class is mainly studied for human L1 element using an assay to monitor retrotransposition in cultured human HeLa cells (16) and SART1 of the silkworm using the Autographa californica nuclear polyhedrosis virus (AcNPV) expression system (17). Although L1 and SART1 both have 3′UTR and poly (A) tail at their 3′ end, they show different requirement for the RNA sequence to initiate the reverse transcription. The presence of 3′ UTR is dispensable for L1 retrotransposition, but poly (A) tract seems essential (16). In addition, active L1 elements possess the ability to transduce non-L1 DNA flanking their 3′ ends to new genomic locations (3,18,19). In contrast, the recognition of 3′UTR by SART1 proteins is crucial for the SART1 retrotransposition (17), and the loss of poly (A) tract resulted in a severe loss of the efficiency for the initiation of reverse transcription (20).
R1Bm is a member of recently-branched subtype, and inserted specifically into the 28S rDNA of the silkworm (9,21). The R1 insertion site is located 74 bp downstream of the R2 site (Figure 1A) (22). R1Bm is flanked by specific 14 bp target site duplication (TSD), and lacks the poly (A) tract at its 3′ end. To understand the basal mechanisms of TPRT, it is of great interest to know how this poly (A)-less R1 initiates its reverse transcription. In this study, we have newly cloned the R1 element from the silkworm genome, and firstly succeeded in retrotransposing the R1 sequence into the 28S rDNA target of the host cells with recombinant baculovirus, AcNPV. Using the in vivo retrotransposition system of R1, we studied the functional roles of 3′UTR of R1 RNA and co-expressed 28S rRNA downstream of R1 sequence.
Figure 1.
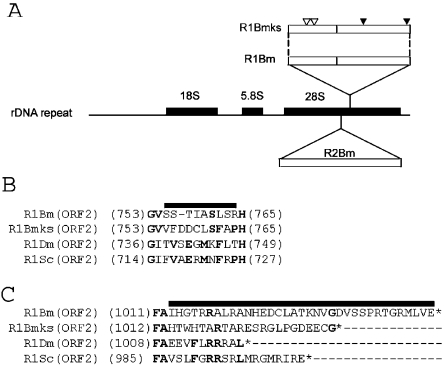
Identification of a new subtype of the R1 element from the silkworm, Bombyx mori. (A) Schematic representation of 28S gene-specific LINEs. Shown in the middle is a diagram of the rDNA unit of Bombyx mori. The newly identified R1 clone (named R1Bmks) is integrated into the sequence 74 bp downstream of the R2 insertion site. Open reading frames (ORFs) are depicted by open boxes. The positions of non-synonymous mutations in the ORF1 and nucleotide insertions in the ORF2 (see B and C) are shown by white and black arrowheads, respectively. (B and C) Altered amino acid sequence of R1Bmks in the ORF2. Insertion of 3 nt within the RT domain results in alteration of 10 amino acids (indicated by bold line), compared with R1Bm (B). Addition of a single nucleotide in R1Bmks alters the amino acid sequences and made the length of ORF2 shorter by 12 amino acids, compared with R1Bm (C). The amino acids conserved between more than two elements are indicated by bold letters. Putative translational stop was indicated by asterisks. R1Bm clones are aligned with R1Dm (X51968; accession number) from D.melanogaster; R1Sc (L00945) from Sciara coprophila.
MATERIALS AND METHODS
Plasmid construction
The R1 ORF1/ORF2/3′UTR portion was directly amplified by PCR with Pfu Turbo DNA polymerase (Stratagene) using primers R1 S519, R1 A5136 (Table 1) and 5 ng of the genomic DNA extracted from Bombyx mori strain, Kinshu × Showa. The reaction mixture was denatured at 94°C for 1 min, followed by 30 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 12 min. The PCR products were loaded on 0.6% agarose gel, and electrophoresed. The DNA band in the calculated size was excised from the gel and purified by the QIAquick Gel Extraction Kit (Qiagen). The recovered product was digested with EcoRI and PstI, subcloned between the EcoRI and PstI sites of the pAcGHLTB plasmid (PharMingen). The resulting plasmid, named R1WT-pAcGHLTB, contained the 64 bp polyhedrin 5′UTR and the GST-X5-(His)6-X29 coding gene fused in-frame with MSEEERE of the R1 ORF1, followed by R1 ORF2/3′UTR and the polyhedrin 3′UTR. Point mutations were introduced into R1WT-pAcGHLTB with primer sets listed in Table 1 using QuickChange™ Mutagenesis Kit (Stratagene). The mutation of each plasmid was confirmed by DNA sequencing. The Δ3′UTR mutant was constructed by self-ligation of the DNA fragment amplified from R1WT-pAcGHLTB with the inverse PCR using 5′-phosphorylated primers, R1 A4992 and pAcGHLTB-S3304. R1WT-pAcGHLTB plasmids with various 3′ tails were constructed as follows. First, 333 bp fragment of 3′ junction between R1 and 28S rDNA was amplified by PCR with a primer set, R1 S4861 and 28S rDNA(+57) using the silkworm genomic DNA as a template and was directly cloned into pGEM-T vector (Promega). Second, PCR reaction was conducted between R1 S4861 and the various primers with PstI sites (Table 1), and the resulting fragments were cloned into the NcoI and PstI sites of R1WT-pAcGHLTB.
Table 1.
List of primers
Underlined letters indicate restriction sites used for subcloning. Mutagenized nucleotides are boxed.
Recombinant AcNPV generation and protein extraction
Sf9 cell was propagated as monolayer cultures at 27°C in TC-100 supplemented with 10% fetal bovine serum (Katakura Co., Nagano, Japan) in the presence of penicillin/streptomycin (Gibco-BRL). The recombinant baculovirus was produced as described previously (17). Briefly, R1WT/mutant-pAcGHLTB plasmid was co-transfected with BaculoGold™ DNA (PharMingen) into the Sf9 cells using the Tfx-20 reagent (Promega). The subsequent virus was plaque-purified and amplified, according to the manufacturer's instructions (PharMingen).
Protein extraction and in vivo retrotransposition assay
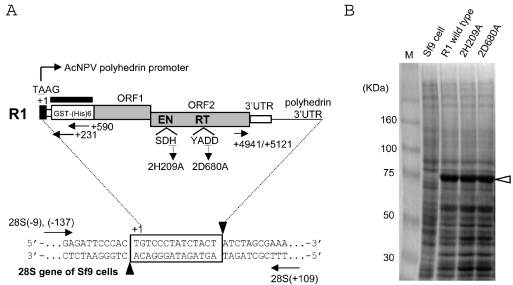
Approximately 5 × 105 of Sf9 cells were infected in a 12-well plate with a R1-containing AcNPV at a multiplicity of 10 plaque forming units (p.f.u.) per cell. At the 12, 24, 48, 72 h post-infection (h.p.i.), the total genomic DNAs from Sf9 cells were extracted with the Puregene DNA isolation kit (Gentra Systems). PCR amplifications were carried out with LA-taq DNA polymerase (TaKaRa), anti-taq antibody (Clontech), ∼2.0 ng of Sf9 genomic DNA, and the primer sets shown below. 3′ junctions of R1 integrations were amplified by the primer sets, +4941/+5121 and 28S (+109). 5′ junctions were amplified using the primer sets, 28S (−9)/(−137) and +231/+590. The reaction mixture was denatured at 96°C for 2 min, followed by 40 cycles of 98°C for 30 s, 62°C for 30 s and 72°C for 30 s. The PCR products were loaded on 2.5% agarose gel, electrophoresed and stained with ethidium bromide. The amplified fragments were directly cloned into the pGEM-T Easy vectors (Promega) and sequenced with an ABI 3100 genetic analyzer (Applied Biosystems). Sequence analyses were conducted with the software, Vector NTI (World Fusion). For the protein analysis, the collected Sf9 cells were washed twice with phosphate-buffered saline (PBS) and subjected to the suspension by the sample buffer containing 50 mM Tris–HCl (pH 6.8), 10% glycerol, 10 mM β-mercaptoethanol and 2% sodium dodecyl sulfate (SDS). The samples were loaded onto the 8% polyacrylamide gel electrophoresis, and the resulting gel was stained with Quick CBB (Wako).
Southern hybridization
5′ junction PCR products were electrophoresed on 2.5% agarose gels and blotted onto nylone membrane (Biodyne B Membrane; PALL BioSupport) in 0.4 N NaOH. After prehybridization, the membrane was hybridized with a ∼550 bp probe corresponding to Glutathione S-transferase (GST) region, which is fused to N-terminus of ORF1 (Figure 2A), at 42°C overnight in 50% formamide, 10× Denhardt's solution (0.2% each of BSA, Ficoll and polyvinylpyrrolidone), 5× SSC, 250 μg/ml salmon sperm DNA and 50 mM NaPO4 (pH 7.0). The probe was labeled with [α-32P]dCTP using Ex Taq polymerase (TaKaRa) by PCR. The primers used for generating the probe are +590 and pAcGHLT-B-S2183.
Figure 2.
In vivo retrotransposition assay for R1 elements using recombinant baculoviruses. (A) Diagram of the PCR assay for retrotransposition. Shown at the top of the figure is a diagram of the R1 element expressed from AcNPV. The 5′UTR sequence derived from the polyhedrin promoter and ORF1/ORF2 of R1 are shaded in black and gray, respectively. Nucleotide position is numbered with the transcription initiation site (A of TAAG) defined as +1. EN and RT denote the endonuclease and reverse transcriptase domains, respectively. The amino acid position of each mis-sense mutant is also shown; the 2H209A mutant represents the substitution of the 209th histidine (H) in the ORF2 for alanine (A), and the 2D680A mutant represents the substitution of the 680th aspartic acid (D) in the ORF2 for alanine (A). Shown at the bottom is a diagram of an rDNA unit showing the location of R1 insertion; 14 bp of the target site duplication (TSD) created upon R1 insertion is boxed. The putative cleavage sites by R1-EN are indicated by black arrowheads. Arrows represent the primer set to amplify the 5′ and 3′ junctions between the R1 sequence and the 28S sequence. The thick bar above the GST region which is fused to the R1 ORF1 indicates the probe used for Southern hybridization in Figure 4. (B) Expression of R1 proteins with a baculovirus-based expression system. The total proteins were extracted from Sf9 cells infected with recombinant viruses shown above and run on SDS–PAGE; the predicted molecular weight for R1 GST/(His)6/ORF1 was 78 170. Lane M; size markers.
RESULTS
Structural features of newly cloned R1Bm
We have newly cloned a complete sequence of R1 (4618 bp) from the genomic DNA of the silkworm, Bombyx mori to develop retrotransposition assay specific for 28S rDNA. As shown in Figure 1A, this clone [named ‘R1Bmks’ (R1Bm clone of the silkworm strain, Kinshu × Showa), accession number; AB182560] contained a synonymous mutation in the position 4857 (GAGGTG→GAGGTA; reading frame is underlined) and two non-synonymous mutations in the position 1318 (CCCGGG→CCCGCG; Gly→Ala) and 1345 (GCTGCC→GCTGTC; Ala→Val) (shown as open arrowheads in Figure 1A), compared with the R1Bm sequence published previously [M19755, (9)]
Additional 3 nt were also inserted in the ORF2 (Figure 1A, solid arrowhead in the middle), resulting in the alteration of the amino acid sequence inside the RT domain (Figure 1B, 754-VSSTIASLSRH-764 → 754-VFDDCLSFAPH-765). These regions, which are located just downstream the seventh highly conserved block between RT domains (6), are not conserved among three insect species (Figure 1B). In R1Bmks clone, 1 nt insertion at the end of ORF2 altered the reading frame (Figure 1A, the right solid arrowhead), which makes the length of ORF2 protein shorter by 12 amino acids (1047 → 1035 amino acids). As shown in Figure 1C, the amino acid sequences and the length of the C-terminal regions of ORF2 are also not conserved among three relatively close insect species. The alterations of amino acid sequence shown above were limited to the non-conserved regions between R1 elements.
R1Bmks retrotransposes into the specific site of 28S rDNA
To figure out whether R1Bmks is active for retrotransposition, we have introduced in vivo R1Bm retrotransposition assay using baculovirus-based expression system (17). We have expressed R1Bmks with its 5′UTR replaced by glutathione S-transferase (GST) and histidine tag, (His)6 under the control of AcNPV polyhedrin promoter in the Sf9 cells. De novo retrotranspositions into genomic DNA of Sf9 cells were detected by 40 cycles of PCR using primers designed to amplify the junction regions between R1 sequence and 28S rDNA sequence (Figure 2A). GST-His-R1Bmks ORF1 fusion protein expressions comprising the molecular size of 78 kDa were confirmed by SDS–PAGE for every R1Bmks recombinant viruses (Figure 2B). The band derived from ORF2 expression in the calculated size, 116 kDa was not observed in wild-type and mutated R1 elements. ORF2 expression is also not observed in other non-LTR retrotransposons such as SART1 in Coomassie staining (unpublished data), and in L1, it is only observed by immunohistochemistry with anti-ORF2 protein antibody (23). However, retrotransposition activity is observed for both SART1 and L1, therefore, it is suggested that even though the quantity is small, enough amount of ORF2 is expressed for retrotransposition.
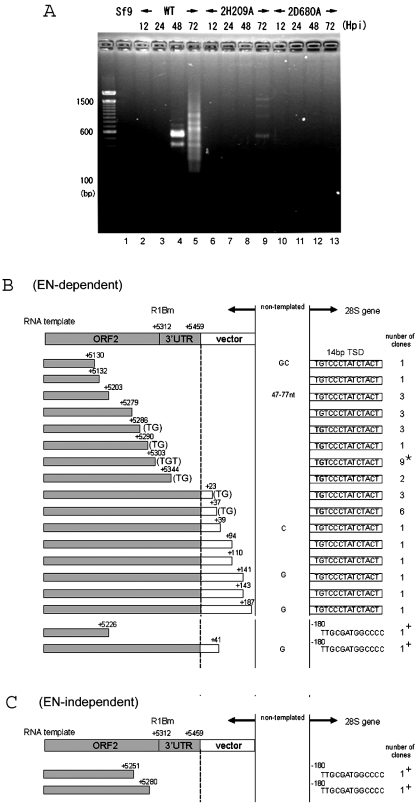
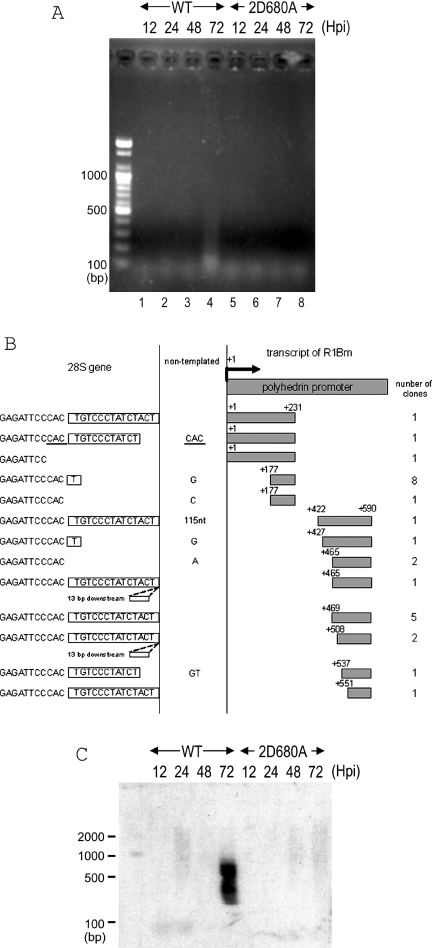
First, the 3′ junctions between the retrotransposed R1 element and the 28S gene were analyzed (Figure 3). A single round of PCR gave rise to the bands indicating the retrotransposition events at 48 and 72 h.p.i. of recombinant AcNPV constructs (Figure 3A). As seen in lane 4, there were only two bands of 400 and 600 bp at 48 h.p.i. At 72 h.p.i., the total intensity and the number of bands increased to the range of 200–1000 bp as in lane 5. In order to know whether these bands represent retrotransposed copies, we have cloned total PCR products into a cloning vector, and sequenced 16 clones for lane 4 (data no shown) and 40 clones for lane 5 (Figure 3B).
Figure 3.
3′-junction analysis for retrotransposed R1 elements. (A) PCR amplification of the 3′ boundaries between the transposed R1 and the 28S rDNA gene. Sf9 genomic DNAs were extracted 12, 24, 48, and 72 h post-infection (h.p.i.) with AcNPV expressing wild-type R1, 2H209A (EN-deficient mutant) and 2D680A (RT-deficient mutant). The purified DNA was used as template for PCR amplification with a pair of primers, +4941 and 28S(+109) (Figure 2A). The PCR products were subjected to 2.5% agarose electrophoresis and stained with ethidium bromide. The molecular size marker is loaded alongside and each size in base pair (bp) was shown on the left of the picture. (B and C) 72 h.p.i. 3′ junction clones obtained with wild-type R1 (B) and endonuclease-deficient R1 (2H209A) (C). Shown at the top of each figure is a diagram of the 3′ end structure of the construct. Sequences derived from the R1Bm and the pAcGHLTB vectors are indicated by shaded and open boxes, respectively. The initiation sites for reverse transcription (left of the dotted vertical lines) are indicated by nucleotide numbers. The target DNA regions are shown on the right of the dotted vertical lines. Extra nucleotides at the junction that are not derived from either the 28S gene or the R1 construct are given between the two vertical lines (non-templated). Boxes to the right of the vertical lines represent the 14 bp of TSD. The number of clones containing each insertion type is indicated in the right-most column and the most major type is indicated by an asterisk. The TGT or TG sequences on the 3′ end of the R1 template that can base-pair with the target DNA are also indicated (Figure 6). +, insertions into the site 180 bp upstream of TSD observed for wild-type and endonuclease-deficient R1.
For 48 h.p.i., 15 clones were integrated into the specific site of 28S rDNA with 14 bp of target-site duplications. Endogenous R1 elements from the silkworm are shown to be flanked by a specific 14 bp TSD of 28S rDNA (9), suggesting that R1Bmks cleaved the specific target site and integrated into the identical position to the endogenous R1 elements. It is of interest that the 5′ end of TSD was adjoined to various sequences of R1; there were 5 clones which adjoined ATACA+5249, 7 clones for AGGAC+5286, 1 clone for GACGG+5361 and 2 clones which adjoined the R1 3′ end.
For 72 h.p.i., 38 out of 40 clones were integrated into the specific site of 28S rDNA with the TSD. There was a wide variety in the R1 sites adjoining the 5′ end of TSD, suggesting that reverse transcription initiated from various positions of template RNA. Some clones contained the downstream AcNPV vector region, implying reverse transcription from read-through transcripts with the non-R1 sequence. It is noteworthy that 24 of 40 clones for 72 h.p.i. and also 13 out of 16 clones for 48 h.p.i. had the identical sequence (TG or TGT, shown within parentheses in Figure 3B) at the 3′ end of the RNA template region (i.e. the start site of reverse transcription). The 5′-UG-3′ or 5′-UGU-3′ sequences are presumably base-paired with the bottom strand of TSD, 5′-AGTAGATAGGGACA-3′, suggesting the interaction between the template RNA and the target 28S rDNA sequence in the initial process of TPRT (see Figure 6). According to this hypothesis, the reverse transcription should start from the position next to the TGT or TG sequences. Among 40 clones, only two clones (Figure 3B; denoted as +) inserted into the sequence other than R1-specific integration site, which is 180 bp upstream of TSD.
Figure 6.
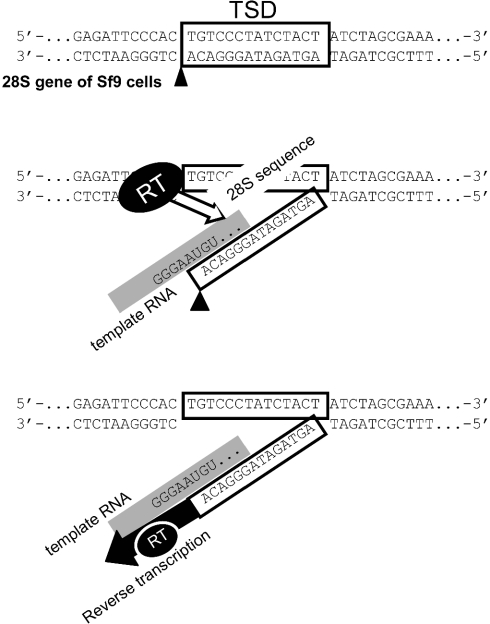
Schematic representation of initial process of TPRT in the R1 element. (Top) First, R1-EN cleaves the non-coding bottom strand of 28S rDNA in the A–C junction. Boxes represent the 14 bp of TSD. (Middle) Then, the target DNA is partially denatured, allowing the UGU on the RNA template to base-pair with the loose target DNA. The template RNA is indicated by a gray line. In this model, the read-through 28S rRNA sequence is base-paired with the DNA target in longer region. During this process, RT of R1Bm may recognize the 3′UTR and base-paired region (open arrow), and place the RNA template at the accurate position for initiation of reverse transcription. (Bottom) Next, reverse transcription starts from the position next to the UGU sequence of template RNA, using the 3′-OH of A residue as primer.
We also generated the recombinant R1Bm constructs with single-amino-acid substitutions at the conserved motif in EN domain and RT domain (Figure 2A). While 2D680A mutation abolished in vivo R1 retrotransposition as shown for several LINEs (Figure 3A, lane 10–13) (16,17), very faint bands were observed for the 2H209A mutant with a non-sense mutation in the catalytic motif of EN domain at 72 h.p.i. (Figure 3A, lane 9). The two recovered fragments from lane 9 indicated the R1 integration into the 180 bp upstream of TSD (Figure 3C), which is identical for two clones of wild-type integration (Figure 3B). These results indicate that a DNA nick has been already pre-formed at −180 of the R1 target site in 28S rDNA, and R1Bmks use this nick for the endonuclease-independent retrotransposition.
5′-junction of integrated R1Bmks elements
PCR amplification of the 5′-junction using the primers complementary to the 28S rDNA and polyhedrin promoter gave rise to faint bands, ranging from 100 to 500 bp (Figure 4A, lane 4). The sizes of these bands were much shorter than expected from the putative full-length product, 590 bp plus the 28S rDNA (Figure 2A). To clarify the figure, we transferred the PCR products to a nylone membrane and performed Southern hybridization with a GST probe (Figure 4C). There were signals in good accordance to full-length products (590 + 137 bp) to 5′ truncated products (>137 bp). To investigate the 5′ junction sequences, we cloned and sequenced the PCR products from Figure 4A, lane 4. All the sequenced clones turned out to be variable 5′-truncated R1 elements connected with the 28S gene sequence (Figure 4B). The variable 5′ truncation was shown to be resulting from a frequent dissociation of RT from the RNA template before completion of the reverse transcription; a diagnostic feature of LINE retrotransposition (24). Some clones contained precise 14 bp TSD. The others, however, showed some aberrations such as deletions in the 28S rDNA gene, non-templated nucleotides addition, and 13 bp insertions of downstream sequence. To detect the full-length R1 insertion, we used another primer complementary to the sequence upstream of +231. One of the resulting fragments (Figure 4B, top) contained the precise junction between TSD and the polyhedrin RNA 5′-terminal sequence (AAG, Figure 2A). One clone included additional CAC nucleotides which was identical to the 28S gene sequence just upstream of TSD (Figure 4B, underlined). Several clones showed aberrations in the 28S gene and/or non-templated nucleotide additions. While these variations shown above were somewhat different from those found in endogenous R1 elements from the silkworm or Drosophila genomic DNA (9,25), retrotransposition of full-length R1 sequence into the specific site indicates the authentic de novo retrotransposition of R1Bmks via its RNA transcript.
Figure 4.
5′-junction analysis for retrotransposed R1 elements. (A) PCR amplification of the boundaries of the transposed R1 5′ ends with the 28S gene. The DNA extraction and PCR analyses were basically same as in Figure 3. A pair of primers, +590 and 28S(−62) was used for amplifying the 5′ junctions. 2D680A, RT-deficient mutant (Figure 2A). (B) 72 h.p.i. 5′ junction obtained with wild-type R1. Shown at the top of the figure is a diagram of the 5′ end structure of R1WT-pAcGHLT. Sequences derived from the pAcGHLTB vector are indicated by shaded boxes. The numbers on the left and the right of the shaded boxes correspond to the nucleotide position numbered with the transcription initiation site. The gray boxes on the right are cDNA and the open boxes on the left are the 28S rDNA regions. Extra nucleotides at the junction that are not derived from either the 28S gene or the R1 construct are given between two vertical lines. Boxes to the left of the vertical lines represent the 14 bp of TSD. Duplicated nucleotides are underlined. Two of the insertions contained 13 bp downstream sequences at the insertion site. The number of clones containing each insertion type is indicated in the right-most column. The top five sequences are from PCR with primer +231, and the bottom eight sequences are from PCR with primer +590. (C) Southern hybridization of the retrotransposed R1 5′ junction PCR. The PCR reaction was conducted by primers +590 and 28S (−137). The PCR products were transferred to a nylon membrane and hybridized with the GST probe (Figure 2A). The DNA size marker was electrophoresed simultaneously and shown on the left.
Sequence in the 3′-terminus affects efficiency and accuracy of retrotransposition
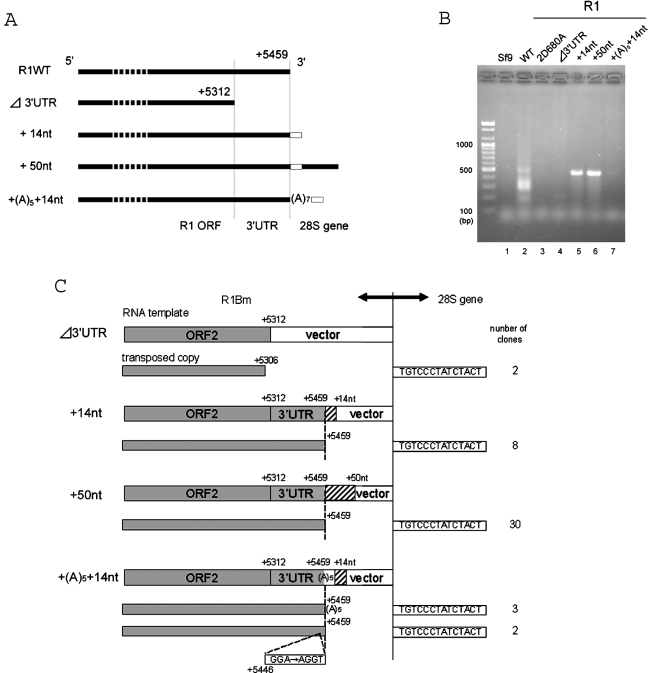
R2 element, 28S rDNA-specific LINE with a single ORF, requires several regions in 3′UTR for initiating the target-primed reverse transcription in vitro (11). To clarify the function of 3′UTR in the R1 retrotransposition, we made an R1 Δ3′UTR-AcNPV construct which lacks the entire R1 3′UTR (Figures 5A and C). Note that this construct retains a downstream polyhedrin 3′UTR. As shown in Figure 5B (Δ3′UTR; lane 4), the band was very faint, but two clones representing the authentic reverse transcription from ORF2 sequence of R1 were recovered from the PCR product (Figure 5C, Δ3′UTR). This result suggests that 3′UTR is not necessary absolutely for the R1 retrotransposition although loss of the sequence lowered severely the retrotransposition efficiency.
Figure 5.
Effects of downstream sequences on the R1 retrotransposition. (A) Diagram of the R1 constructs with various 3′ end structures; 14 and 50 nt of the downstream 28S gene are added to the R1WT construct. The 14 bp sequence of TSD is indicated by an open box. Note that the 3′ termini of each construct is followed by the AcNPV-derived polyhedrin 3′UTR. Transcription start, +1. (B) PCR amplification of the 3′ boundaries between the transposed R1 copies and the 28S gene. DNA extraction and PCR reaction are basically same as in Figure 3. The primer set used for PCR was +5121 and 28S(+109). 2D680A, RT-deficient mutant. (C) 72 h.p.i. 3′ junctions obtained with various R1 construct in (A). Shown at the top of each figure is a diagram of the 3′ end structure of the constructs. Sequences derived from R1Bm and additional 28S downstream sequences are indicated by shaded and hatched boxes, respectively. The vertical dotted lines represent the boundary between the cDNA region and the target DNA region. The initiation site for reverse transcription (left of the dotted vertical lines) is indicated by the numbers. Boxes to the right of the vertical lines represent the 14 bp of TSD. The number of clones containing each insertion type is indicated in the right-most column. Two clones amplified from the + (A)5 + 14 nt construct contained mutations at the 3′ end of 3′UTR (G of GAA corresponds to the position +5446).
As shown above, the short UG or UGU sequences in RNA of WT R1 are potentially base-paired with the target DNA during initial process of TPRT (refer to Figure 6). Then, we next tested whether the long base-pairing between template RNA and target DNA raises the efficiency or accuracy of R1 retrotransposition. We made recombinant baculoviruses with various length of 28S rDNA sequence located downstream of R1 elements (Figure 5A, +14 nt, +50 nt, and + (A)5 + 14 nt). In striking contrast to R1WT, addition of downstream 14 and 50 nt from 28S rDNA showed a strong single band representing the retrotransposition (Figure 5B, lanes 5 and 6). All the sequenced clones (38 clones in total) from lanes 5 and 6 showed that the transposed copies have the precise junction between the 3′ end of 3′UTR of R1 and the 28S target sequence as in the endogenous R1Bm elements (Figure 5C, +14 nt, +50 nt). These findings suggest that the downstream 28S sequence greatly enhanced the accurate initiation of reverse transcription from the EN-digested site.
Site-specific LINEs usually have poly (A) tract at their 3′ tail, but R1Bmks contains only two A nucleotides at its 3′-terminus (data not shown). To clarify the functional role of the 3′-terminal sequence of R1, five A nucleotides (A5) were added to the +14 nt construct between the R1-28S junction (Figure 5A). As shown in Figure 5B, the retrotransposition efficiency was drastically reduced, compared with the +14 nt construct, indicating that addition of poly (A) tail resulted in a severe defect in retrotransposition even if the poly (A) tract was followed by downstream 28S sequence. Among 5 sequenced clones, three showed the reverse transcription from the poly (A) tract. Two clones showed the reverse transcription from +5459 and some aberrations inside the R1 sequence.
The above results indicate that R1Bm transcription pass through into the 28S rDNA region in vivo, that transcripts are base-paired strongly with the target DNA sequence, and that the minimal interval between the base-paired region and the predestinate RT start site is important for efficient and accurate retrotransposition of R1 (Figure 6).
DISCUSSION
The newly cloned R1 from the silkworm genome was found to be active for retrotransposition, and it turned out that the baculovirus-based in vivo retrotransposition assay could be applied to the study of various site-specific LINEs (17,26). The most distinctive feature of R1 retrotransposition is its requirement of 3′ downstream sequence derived from 28S gene for the precise, efficient integration into its target DNA. Luan and Eickbush reported that addition of downstream target sequence to the R2 element resulted in the accurate start of reverse transcription from the precise R2-28S gene junction (10,14). The R2 insertion site is located just 74 bp upstream of the R1 site (9), but R1 and R2 are distantly related from the phylogenetic study (6). It is of some interest that these distantly related LINEs showed the similar requirements of downstream 28S sequence for the precise integration.
Phylogenetically, R1 element belongs to the ‘R1 clade’ (6). Most R1 clade non-LTR retrotransposons are target-specific and show the diversified target specificities (27,28), suggesting that they have evolved to change their target sequences from the common ancestors. Using in vivo retrotransposition assay, we previously characterized another R1 clade element, SART1, which integrates into the telomeric repeats (TTAGG)n (17,20). The comparison between R1 and SART1 revealed some distinct features in the initial processes of reverse transcription. The full-length SART1 element with poly (A) at the 3′ terminal initiates its reverse transcription from the poly (A) tract (17). However, the complete R1Bm unit with 3′UTR started its reverse transcription from various positions of template RNA (Figure 3B), suggesting that reverse transcriptase of R1 recognizes its 3′UTR in a less stringent manner than that of SART1. SART1 was shown to recognize some motifs within the 3′UTR and the loss of the 3′ terminal poly (A) tract severely decreased its efficiency for integration (20), while R1 elements do not contain A-rich tract at its 3′ junctions.
As shown in Figure 5, the loss of 3′UTR decreased the retrotransposition activity of R1Bm, compared to WT construct with 3′UTR. This indicates that RT encoded in ORF2 of R1Bm also recognizes some structure in 3′UTR, but it seems insufficient for specifying the start position of reverse transcription. In case of WT R1Bm retrotransposition, many copies started reverse transcription from UG or UGU (Figure 3B), which are presumably base-paired with the 3′ end of the bottom strand of target DNA (Figure 6). The similar short base-paring between the template RNA and the target DNA was also presumed in the SART1 element, which lacks 3′-half of 3′UTR or poly (A) tract (20). Thus, the short interaction between RNA and DNA seem to play some roles in promoting the initial process of TPRT in R1 and SART1. To ensure the accurate and effective retrotransposition, R1Bm needed the downstream 28S sequences in addition to 3′UTR, such as 3′-half of 3′UTR or poly (A) in SART1 element. In this context, the read-through 28S rRNA sequence of R1Bm may enable the long base-paring between RNA and DNA, and contribute to increase the accuracy for reverse transcription and the retrotransposition efficiency (Figure 5B). R1 elements are oriented in the same direction as the ribosomal gene transcription, and in Drosophila melanogaster, endogenous R1 transcripts include upstream and downstream 28S sequences (12), so it is suggested that R1Bm is co-transcribed with the 28S genes. We suggest that constructs most reflecting the endogenous R1Bm are +14 nt/+50 nt and the annealing system between read-through RNA and the target DNA may be actually used in endogenous R1Bm. It is of some interest that addition of 5 nt poly (A) tract at the end of R1Bm resulted in a severe loss of retrotransposition activity (Figure 5B), suggesting that the spatial distance between the R1 3′UTR and the downstream 28S rRNA sequence is somehow recognized by RT of R1Bm (Figure 6). The spatial arrangement might be disturbed by insertion of 5 nt of A tract.
Retrotransposition by R1 also showed the characteristic feature of LINE at the 5′ boundary; frequent 5′ truncation (Figure 4B), due to the incomplete reverse transcription of RNA template into cDNA. Some 5′ border of endogenous R1Bm contains duplication of R1 sequence, but no 28S sequence deletion or 5′ truncations are found (9). However, R1Bm construct in this assay showed various 5′ junction structure and severe aberrations, such as the deletion of 28S sequence by 17 bp (Figure 4). This may be due to the replacement of upstream 28S sequences and R1-5′UTR to polyhedrin 5′UTR and GST sequence. The inclusion of 28S sequence to its 5′ termini of the construct achieved intact 5′ junction formation in R2 (26,29), so it may be the same for R1. This implicates that the free ends generated during this process are repaired by the cellular proteins.
It was unexpected that the retrotransposition into the site 180 bp upstream of R1 target site was observed for R1 2H209A mutant, which contains a non-sense mutation in the catalytic motif of EN (Figure 3A). This mutation has been shown to severely decrease its endonucleolytic activity of R1-EN in vitro (Aoyagi and Fujiwara, unpublished data), and the primary sequence around the hotspot (−180) showed no similarity with that of the specific cleavage site by R1-EN in vitro (15), indicating that this integration was not resulted from the intrinsic EN activity of R1Bm. The position −180 was only site for integration other than the R1-intrinsic target (four clones in Figure 3B and C) and their reverse transcription started various regions of template RNA, suggesting that the −180 position was precedently formed in 28S rDNA before the EN of R1Bm works during TPRT reaction. A low level of retrotransposition by human L1 which lacks its endonuclease activity was reported in vivo and in vitro (31,32). This suggests that such L1 integrations occurred at the pre-formed double-strand break on the chromosomal DNA. Although the mechanism to generate a nick on this hot spot (−180) was unclear, the DNA cleavage might be pre-formed possibly by the recombination event around this spot.
Acknowledgments
We thank K.K. Kojima for critical reading of the manuscript. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (MESCJ) and by a Grant-in-Aid from the Research for the Future Program of the Japan Society for the Promotion Science (JSPS). T.A. and M.O. are recipients of Research fellowships of the JSPS for Young Scientists. Funding to pay the Open Access publication charges for this article was provided by MESCJ.
Conflict of interest statement. None declared.
REFERENCES
- 1.Arkhipova I., Meselsonm M. Transposable elements in sexual and ancient asexual taxa. Proc. Natl Acad. Sci. USA. 2000;97:14473–14477. doi: 10.1073/pnas.97.26.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Moran J.V., DeBerardinis R.J., Kazazian H.H., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 5.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 6.Malik H.S., Burke W.D., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 7.Burke W.D., Calalang C.C., Eickbush T.H. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol. Cell Biol. 1987;7:2221–2230. doi: 10.1128/mcb.7.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Malik H.S., Eickbush T.H. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl Acad. Sci. USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong Y., Eickbush T.H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol. Cell Biol. 1988;8:114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan D.D., Eickbush T.H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 12.Long E.O., Dawid I.B. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979;18:1185–1196. doi: 10.1016/0092-8674(79)90231-9. [DOI] [PubMed] [Google Scholar]
- 13.Kidd S.J., Glover D.M. Drosophila melanogaster ribosomal DNA containing type II insertions is variably transcribed in different strains and tissues. J. Mol. Biol. 1981;151:645–662. doi: 10.1016/0022-2836(81)90428-9. [DOI] [PubMed] [Google Scholar]
- 14.Luan D.D., Eickbush T.H. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Q., Schumann G., Boeke J.D. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl Acad. Sci. USA. 1998;95:2083–2088. doi: 10.1073/pnas.95.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi H., Fujiwara H. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 2002;21:408–417. doi: 10.1093/emboj/21.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickeral O.K., Makalowski W., Boguski M.S., Boeke J.D. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodier J.L., Ostertag E.M., Kazazian H.H., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 20.Osanai M., Takahashi H., Kojima K.K., Hamada M., Fujiwara H. Essential motifs in the 3′ untranslated region required for retrotransposition and the precise start of reverse transcription in non-long-terminal-repeat retrotransposan SART1. Mol. Cell Biol. 2004;24:7902–7913. doi: 10.1128/MCB.24.18.7902-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubczak J.L., Burke W.D., Eickbush T.H. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl Acad. Sci. USA. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara H., Ogura T., Takada N., Miyajima N., Ishikawa H., Maekawa H. Introns and their flanking sequences of Bombyx mori rDNA. Nucleic Acids Res. 1984;12:6861–6869. doi: 10.1093/nar/12.17.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ergun S., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 24.Ostertag E.M., Kazazian H.H., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Gonzalez C.E., Eickbush T.H. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics. 2002;162:799–811. doi: 10.1093/genetics/162.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto H., Hirukawa Y., Tani H., Matsuura Y., Hashido K., Tsuchida K., Takada N., Kobayashi M., Maekawa H. Integration of the 5′ end of the retrotransposon, R2Bm, can be complemented by homologous recombination. Nucleic Acids Res. 2004;32:1555–1565. doi: 10.1093/nar/gkh304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima K.K., Fujiwara H. Evolution of target specificity in R1 clade non-LTR retrotransposons. Mol. Biol. Evol. 2003;20:351–361. doi: 10.1093/molbev/msg031. [DOI] [PubMed] [Google Scholar]
- 28.Anzai T., Takahashi H., Fujiwara H. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell Biol. 2001;21:100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eickbush D.G., Luan D.D., Eickbush T.H. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol. Cell Biol. 2000;20:213–223. doi: 10.1128/mcb.20.1.213-223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 31.Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 32.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]