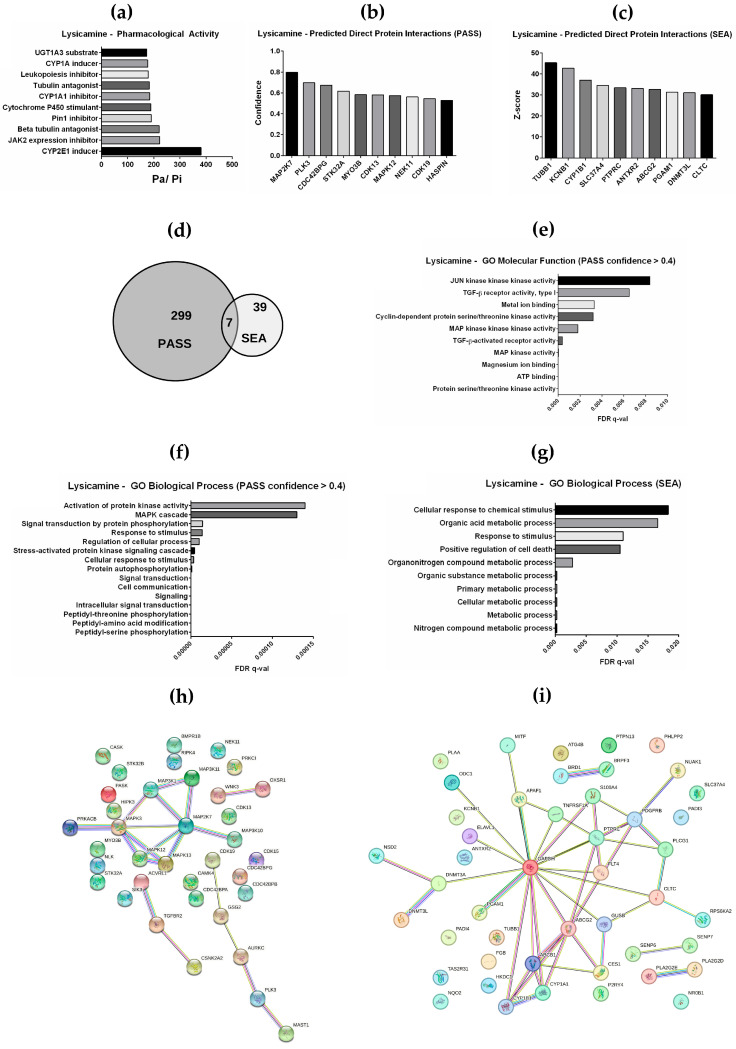
Figure 5.
Lysicamine activity was characterized via in silico analysis. (a) PASS analysis resulted in a list of pharmacological activities. (b) PASS analysis also predicted 36 protein interactions (confidence value > 0.4). (c) SEA analysis predicted 46 direct protein interactions. (d) Among both lists, 7 proteins were shared. Go Biological Process analysis modeled (e) molecular functions and (f) biological processes for proteins predicted by PASS and (g) biological processes by SEA. STRING database was used to generate a protein–protein interaction network for proteins predicted (h) by PASS and (i) by SEA.