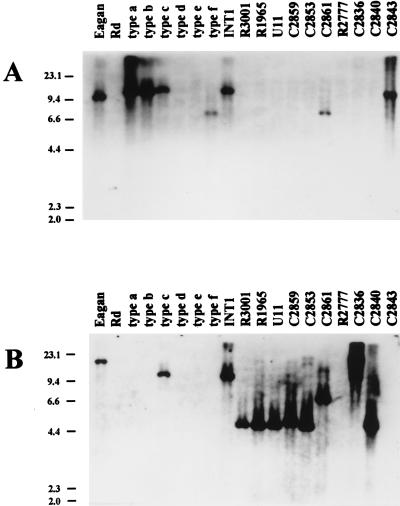
FIG. 6.
Genomic blots with hif- and hic-specific probes. Genomic DNAs were digested with BglII, blotted, and probed as indicated. Assuming conservation of flanking sequence, RFLPs can be predicted from Rd and the purE-pepN sequence by adding 3,700 bp to the distance between purE and pepN translational start sites. Predicted RFLPs are 3,860 bp for Rd (class I), 10,875 for Hib AM30 (class V), 4,503 bp for R3001 (class IIa), 4,749 bp for C2859 (class IIb), and 6,984 bp for C2861 (class IIIa). The positions and lengths (in kilobases) of λHindIII marker fragments are indicated on the left. (A) Blot with the hif probe. Fragment lengths, estimated from mobilities, are 10 to 11 kb for Hib Eagan, Hib C2843, INT1, and reference strains of type a, type b, and type c and 7.5 kb for C2861 and the type f reference strain. (B) The same blot stripped and reprobed with the hic probe. Fragment lengths, estimated from mobilities, are >22 kb for Hib Eagan, >22 kb for C2836, 10 to 11 kb for reference type c and INT1, 7.5 kb for C2861, 5 kb for C2859 and C2840, and 4.4 kb for R3001, R1965, U11, and C2853. C2840 had two slightly resolved fragments of similar size, suggesting a duplication of hic DNA, and C2836 and Hib Eagan have RFLPs for BglII.