Figure 2.
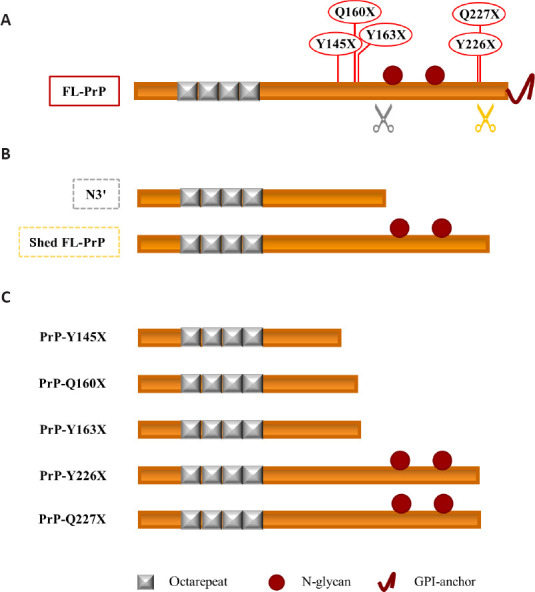
Soluble PrPC proteoforms and pathogenic truncated PrP: a schematic comparison.
(A) Linear representation of PrPC (23–230) showing (i) the octarepeat region (repeated grey boxes), (ii) the N-glycosylation sites (a. a. 181 and 197, red spheres), and (iii) the GPI-anchor (red curved line, extreme C-terminus). Grey and orange scissors indicate γ-cleavage and shedding of PrPC. The pathogenic stop-codon mutations (Y145X, Q160X, Y163X, Y226X, and Q227X) that generate truncated PrPs are shown in red above the linear representation of PrPC. (B) Linear representation of the physiological soluble PrP proteoforms N3’ (non-glycosylated PrP fragment generated by the γ-like-processing that occurs before the N-glycosylation site) and shed FL-PrPC (fully glycosylated and produced by shedding). (C) Linear representation of the pathological truncated PrPs. Truncated Y145X, Q160X, and Y163X PrPs resemble the physiological N-terminal fragment derived by the γ-cleavage-like processing, while truncated Y226X and Q227X PrPs look like the physiological shed PrPC proteoform. Created with Microsoft PowerPoint. FL-PrP: Full-length PrP; GPI: glycosylphosphatidylinositol; PrP: prion protein; PrP-Q160X: truncated Q160X PrP; PrP-Q227X: truncated Q227X PrP; PrP-Y145X: truncated Y145X PrP; PrP-Y163X: truncated Y163X PrP; PrP-Y226X: truncated Y226X PrP.
