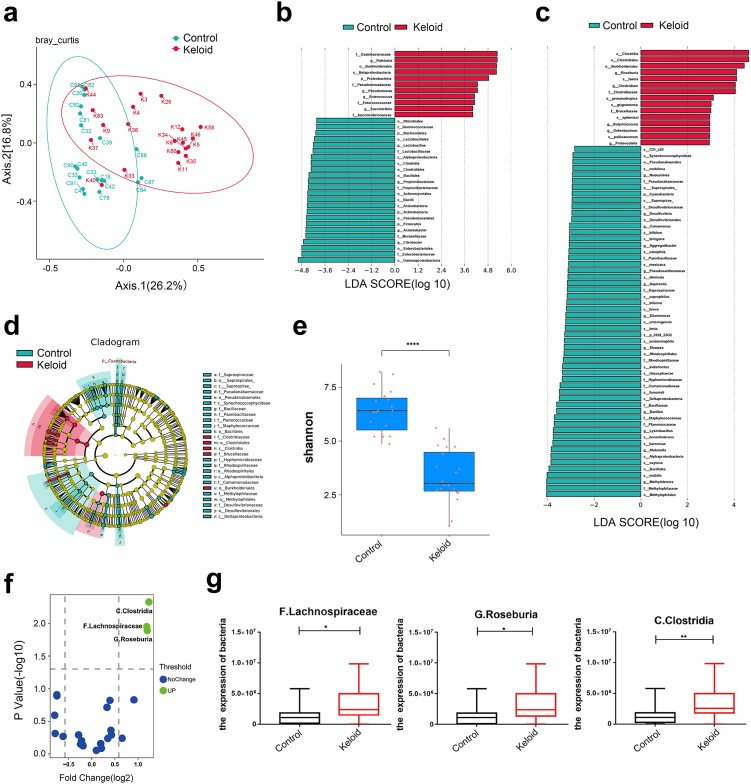
Fig. 1.
Microbiomic analysis of keloid. (a) Principal co-ordinates analysis. The points in the principal coordinate analysis diagram represent the samples, and the distance between the points indicates the degree of difference in the bacterial flora of the samples. The higher the similarity of the microbial community structure was, the smaller the difference. Keloid: 20 keloid tissues. Control: 19 normal tissues. (b and c) From the genus and species levels, each horizontal column represents a species, the length of the column corresponds to the linear discriminant analysis (LDA) value, and the higher the LDA value is, the greater the difference. The color of the column corresponds to which group of characteristic microorganisms the species belongs, and the characteristic microorganisms indicate that the abundance in the corresponding group is high. (d) From the inside to the outside, the cladogram map corresponds to the different classification levels of the genus and family, and the lines between the levels represent the affiliation. Each circle node represents a species; if the node is yellow, it means that the difference between the groups is not significant, and if the node is not yellow, it means that the species is a characteristic microorganism of the corresponding color group. The colored fan-shaped area marks the subclassification interval of the characteristic microorganism. (e) Box plot of the Shannon index of control and keloid. Wilcox Test was used. (f) Volcano map analysis of differentially expressed bacteria by metaproteomics. P < 0.05, Fold change (FC) threshold >1.5. (g) Detection of the expression levels of C. Clostridia, F. Lachnospiraceae and G. Roseburia by metaproteome. Keloid (n = 18), Control (n = 19). Unpaired t test was used. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.