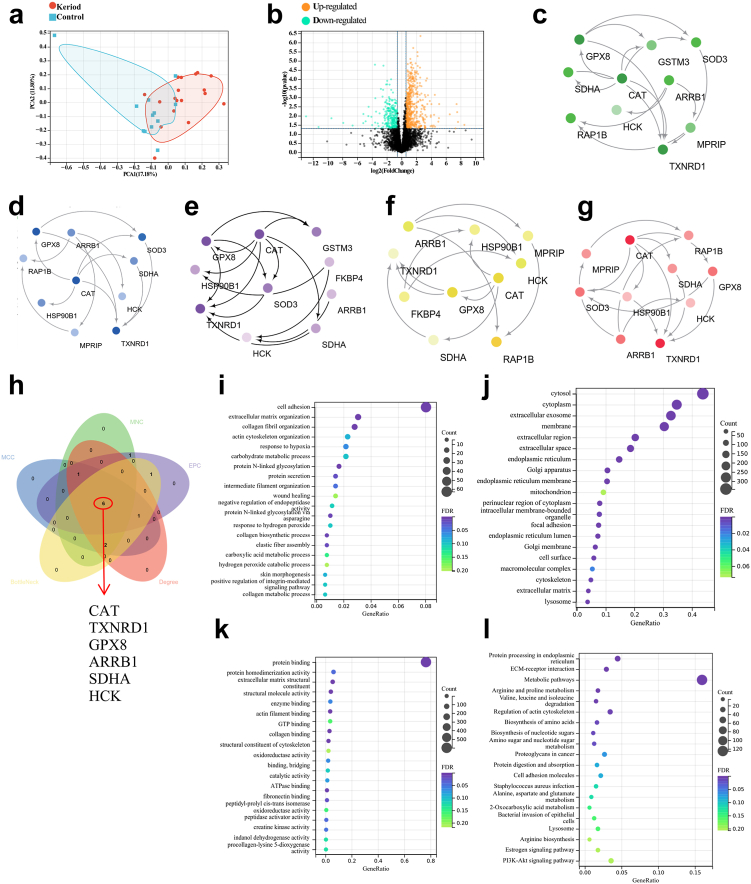
Fig. 4.
Metaproteomics analysis of keloid samples. (a) The abscissa PC1 and ordinate PC2 in the figure represent the scores of the first and second principal components, respectively, and the scatter color represents the sample grouping. 30 samples (18 keloid tissue vs 12 normal skin tissue) were used for metaproteomics. (b) Differential Protein Volcano Plot. The horizontal axis represents the fold change (log2 value) of the DEPs, the vertical axis represents the P value (-log10 value), black represents the proteins with no significant difference, red represents the upregulated proteins, and green represents the downregulated proteins. P < 0.05, Fold change (FC) threshold >1.5. (c–g) Five algorithms of MNC, MCC, EPC, BottleNeck, and degree were used to screen hub proteins through Cytoscape software. (h) Venn diagrams for MNC, MCC, EPC, BottleNeck, and degree algorithms. (i–k) Gene Ontology (GO) analysis (biological process, cellular component and molecular function). (l) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.