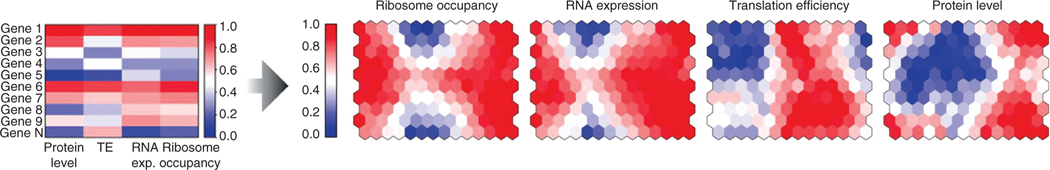
Figure 2.
RNA abundance and protein abundance both correlate better with ribosome occupancy than they do with each other. Expression analysis was performed on lymphoblastoid cell lines of diverse genetic backgrounds taken from the HapMap project. Genes were clustered into modules or neurons (hexagon, right panels) within a self-organizing map based on similar expression profiles across four different measurements (protein abundance, translation efficiency, RNA abundance, and ribosome occupancy; left panel). The right panel displays the same self-organizing map colored to portray the mean expression of the genes within the module based on the four different datasets. The authors ask if hexagons with similar mean expression by one measurement (either both colored red or both colored blue) also show similar expression when using an alternate measure of expression. Ribosome occupancy correlates with RNA expression and protein level better than RNA expression and protein level correlate with each other. Note, ribosome occupancy is defined by the total read counts for an RNA after ribosome profiling, while translation efficiency takes into account the total pool of RNA (RNA-seq) in addition to the ribosome occupancy [reprinted with permission (23)].