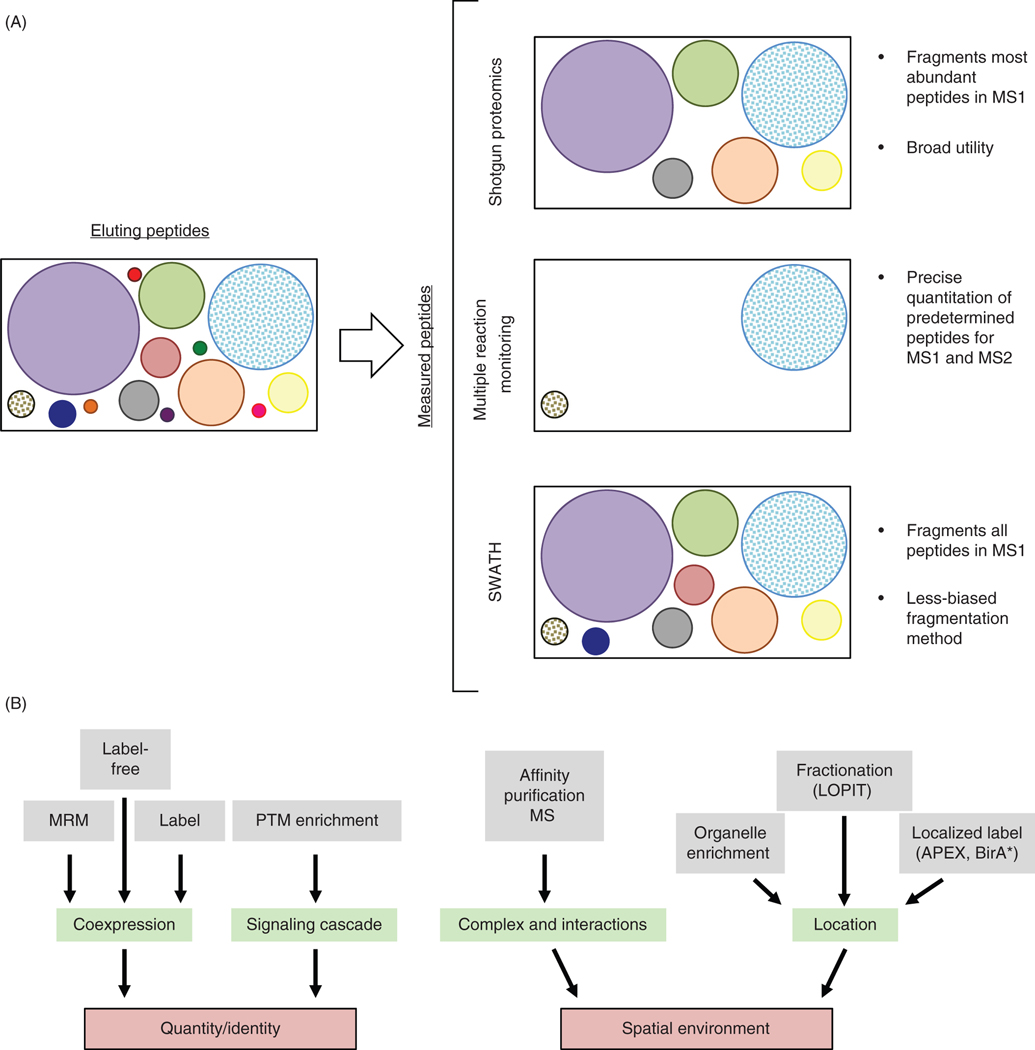
Figure 4.
Mass spectrometry techniques for building protein networks. (A) Peptides (circles; size indicates relative abundance) elute from the LC column into the mass spectrometer. In shotgun/bottom-up proteomics, peptides are scanned in the MS1 and the most abundant ions selected for fragmentation and identification via multiple MS2 scans. In MRM, both the MS1 and MS2 scan are performed on predetermined m/z ratios set by the user to precisely quantify peptides of interest, including low abundant peptides. SWATH by contrast fragments all ions from the MS1 scan, resulting in many more MS2 scans, each containing spectra from many parent ions. (B) Upstream techniques can be used in conjunction with mass spectrometry to enable protein and PTM identification, quantitation, and spatial localization information used to build protein networks.