Figure 5.
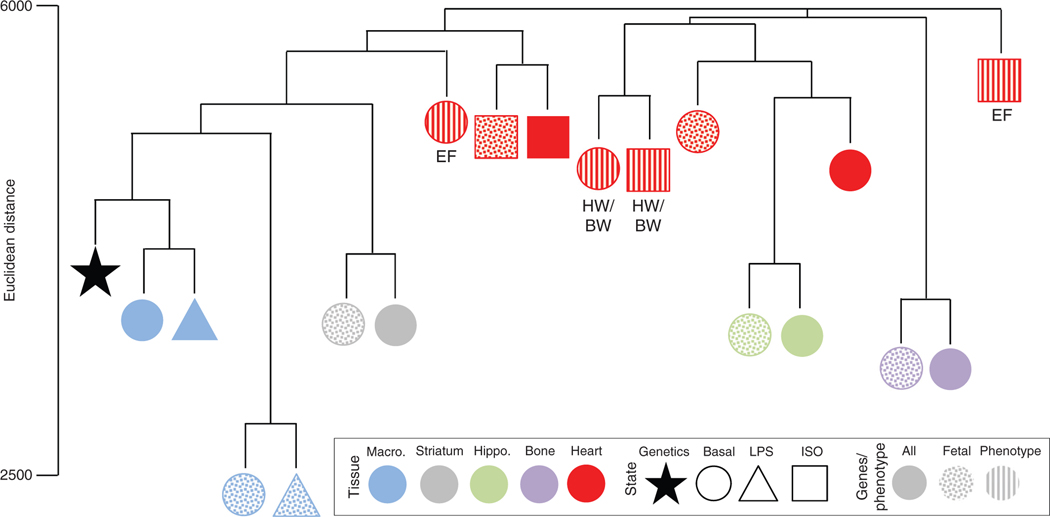
The role of genetics in gene expression is organ specific. To test the relationship between genetics, gene expression, and phenotype, we examined data from a panel of 37 genetically diverse, inbred mouse strains with microarray data from multiple organs: Macrophages with and without LPS stimulation (unpublished), striatum (120), hippocampus (120), bone marrow (38), and heart with and without isoproterenol (ISO) stimulation to induce heart failure (128). Strains were clustered based on expression of all genes on the microarray (All) or a class of genes known as the “fetal gene program” (Fetal), whose cardiac expression are considered to be biomarkers of heart failure. The relatedness between each strain-by-strain comparison (Euclidean) was compared across organs. If the relative similarity in expression between two strains is similar across two organs, those two organs cluster closer together on the dendrogram. We also incorporated genetic relatedness based on kinship matrix derived from SNPs (Genetics). Macrophages cluster according to genetics, suggesting that strains with similar genetics also show similar expression patterns in macrophages regardless of if we examine all genes, or the cardiac fetal genes, and even when examining expression after LPS stimulation. By contrast, other organs, such as bone marrow, have expression relationships that less closely match genetic relationships. For context, we compared the relationships between genetics versus mRNA expression to that of genetics versus cardiac phenotype [ejection fraction (EF) and heart weight/body weight (HW/BW), two indices which change in heart failure]. In some cases, the genetic relationship more closely matched the phenotype than the expression (basal EF), but in other cases it did not (EF after ISO). We hypothesized that the “fetal gene program” was an intermediate between genetics and phenotype, but found that it no more closely matched the phenotypic relationships than when we examined all genes together. These analyses indicate that the relationship between genetic variation, mRNA expression, and ultimately phenotype is buffered at each level. For example, complex SNP interactions and chromatin features may buffer the relationship between genetic variation and mRNA expression, while posttranscriptional and posttranslational processing as well as compartmentalization may buffer the relationship between mRNA and protein levels, with the relationship between protein and phenotype in turn buffered by protein network properties and interaction with other classes of molecules.