Extended Data Fig. 7 |. Recurrent inversions identified with Strand-seq.
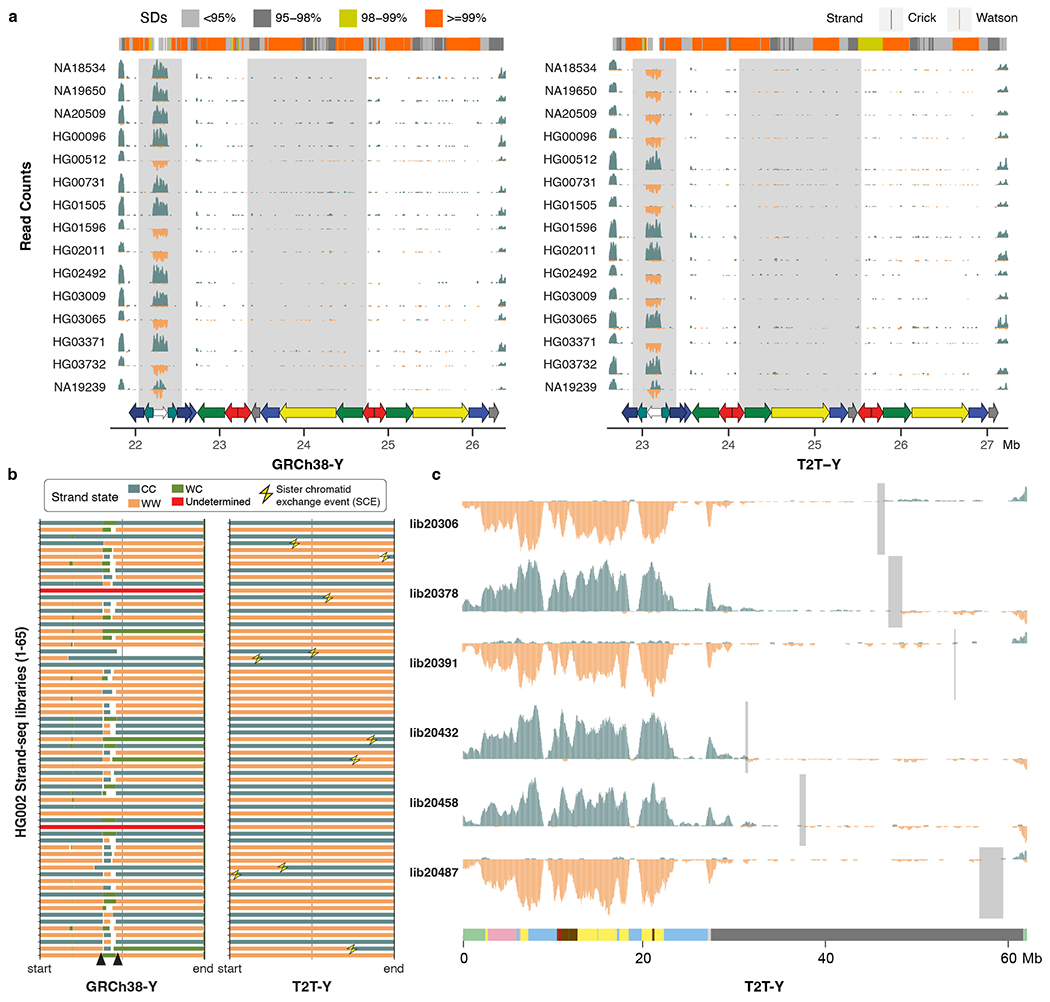
a. Five out of 15 individuals have the inverted variant as present in HG002 at the P3 palindrome (white arrow). Although inversions across P1–P2 (yellow and red arrows) are difficult to confirm with Strand-seq because of the high sequence similarity between the palindromic arms, different orientations are observable in these samples. b. Strand states for 65 Strand-seq libraries of HG002. Depending on the mappings of directional Strand-seq reads (+ reads: ‘Crick’, C, - reads: ‘Watson’, W), reference sequence was assigned in three states: WC, WW, and CC. WC, roughly equal mixture of plus and minus reads; WW, all reads mapped in minus orientation; CC, all reads mapped in plus orientation. Changes in strand state along a single chromosome are normally caused by a double-strand-break (DSBs) that occurred during DNA replication159 in a random fashion and we refer to them as sister-chromatid-exchanges (SCEs, yellow thunderbolts). Recurrent change in strand state over the same region in multiple Strand-seq cells indicates misassembly. Similarly, collapsed or incomplete assembly of a certain genomic region will result in a recurrent strand state change as observed for GRCh38-Y (black arrowheads). In contrast, T2T-Y shows strand state changes randomly distributed along each Strand-seq library with no evidence of misassembly or collapse. c. Strand-seq profile of selected libraries over T2T-Y summarized in bins (bin size: 500 kb, step size: 50 kb). Teal, Crick read counts; orange, Watson read counts. As ChrY is haploid, reads are expected to map only in Watson or Crick orientation. Light gray rectangles highlight regions where SCEs were detected in the heterochromatic Yq12 despite a lower coverage of Strand-seq reads. A modified breakpointR parameter was used (windowsize = 500000 minReads = 20) in order to refine detected SCEs presented in panel b and c.
