Fig. 6 |. Short-read mappability and variant calling improvements on T2T-Y.
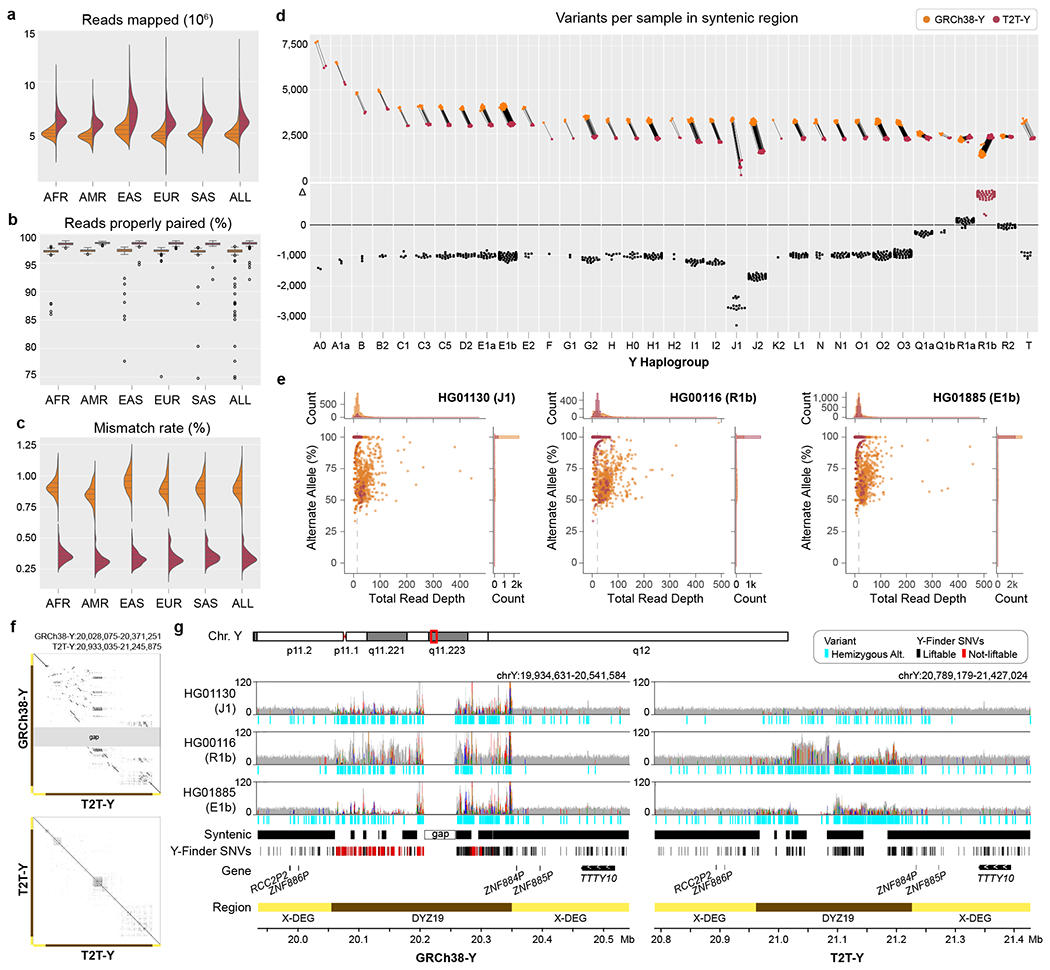
In all plots, GRCh38-Y is colored orange and T2T-Y is maroon. The complete sequence of T2T-Y improves short-read alignment of the 1KGP dataset by a. increased number of reads mapped, b. higher portion of reads properly paired, and c. lower mismatch rate compared to GRCh38-Y. Bar in the box plot represents the 1st, 2nd (median), and 3rd quartile of the data. Whiskers are bound to the 1.5 × interquartile range. Data outside of the whisker ranges are shown as dots. d. The number of called variants within syntenic regions is reduced on T2T-Y for all haplogroups except R1 (haplogroup of GRCh38-Y). e. Further investigation on 3 samples (J1, R1b, and E1b) shows a higher number of variants called with excessive read depth and variable alternate allele fractions for GRCh38-Y. Each dot represents a variant, with the % alternate alleles as a function of total read depth. Dotted line represents the median coverage on T2T-Y, close to the expected 1-copy coverage. f. Dotplot of the DYZ19 array between GRCh38-Y and T2T-Y and self-dotplot of T2T-Y. Large rearrangements are observed, with multiple inversions proximal to the gap in GRCh38-Y with respect to T2T-Y (top), while more identical, tandem duplications are visible in T2T-Y (bottom). g. Read pile-ups and variants on DYZ19 for GRCh38-Y (left) and T2T-Y (right) as shown with IGV71. Gray histogram shows the mapped read coverage, with colored lines indicating non-reference bases with >60% allele frequency. Regardless of the haplogroup, the incomplete DYZ19 array in GRCh38-Y hinders interpretation. Syntenic regions between the two Ys are marked, and SNV sites used to identify Y haplogroup lineages in Y-Finder are shown below, with variants liftable from GRCh38-Y to T2T-Y in black, not-liftable in red, respectively.
