Extended Data Fig. 2 |. Validation and polishing of the T2T-Y.
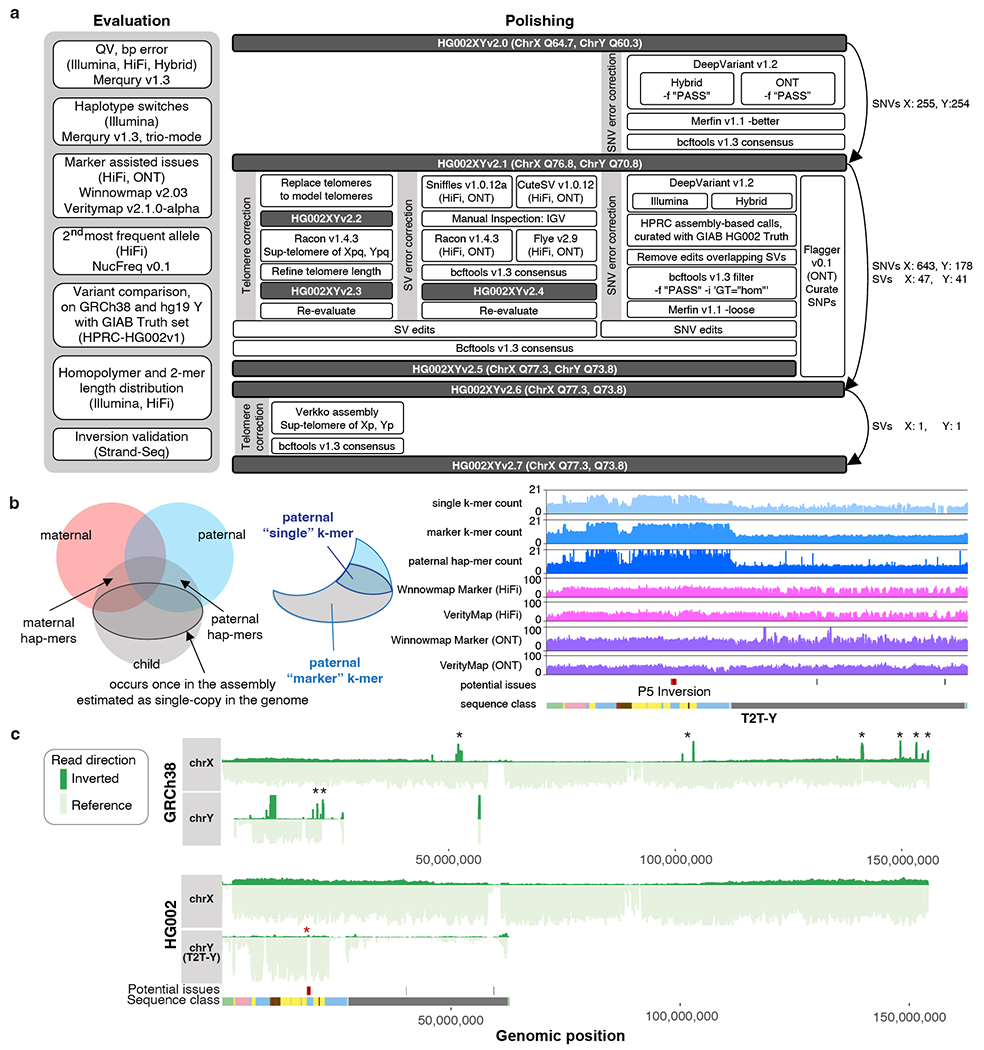
a. Evaluation and polishing workflow performed on T2T-CHM13v1.1 autosomes + HG002 XY assemblies. b. Venn diagram of the k-mers from the parents and child. On the left, hap-mers18 represent haplotype specific k-mers inherited by the child. The darker outlined circle inside the child k-mers represent single-copy k-mers (k-mers occurring once in the assembly and single-copy in the child’s genome). Right figure shows an example of the paternal specific, “single-copy” and “marker” k-mers. The marker set includes both multi-copy and single-copy k-mers specific to the paternal haplotype that were inherited by the child. Unlike polishing the nearly haploid CHM13 assembly17, both single-copy k-mers and marker k-mers were used for the marker-assisted alignments to HG002 XY. This helped align more reads within repetitive regions to the correct chromosome for evaluation during polishing. Right panel shows counts of the k-mers and coverage of HiFi and ONT reads using the marker-assisted Winnowmap2 alignment, in addition to alignments from VerityMap, which uses locally unique k-mers for anchoring the reads. c. Aggregated Strand-seq coverage profile across all 65 libraries on GRCh38-Y (top) and T2T-Y (bottom). Each bar represents read counts in every 20 kb bin supporting the reference in forward direction (light green) or reverse direction (dark green). Multiple spikes in reverse direction (black asterisks) in GRCh38-Y indicate inversion polymorphisms relative to HG002, likely due to differences between the haplogroups. Such spikes in coverage are not observed on T2T-X and T2T-Y, which confirm the structural and directional accuracy of the HG002 assemblies. A 3 kb inversion of the unique sequence between the P5 palindromic arms was identified as erroneous in T2T-Y (red asterisk), but was confirmed to be polymorphic in the population and left uncorrected in this version of the assembly.
