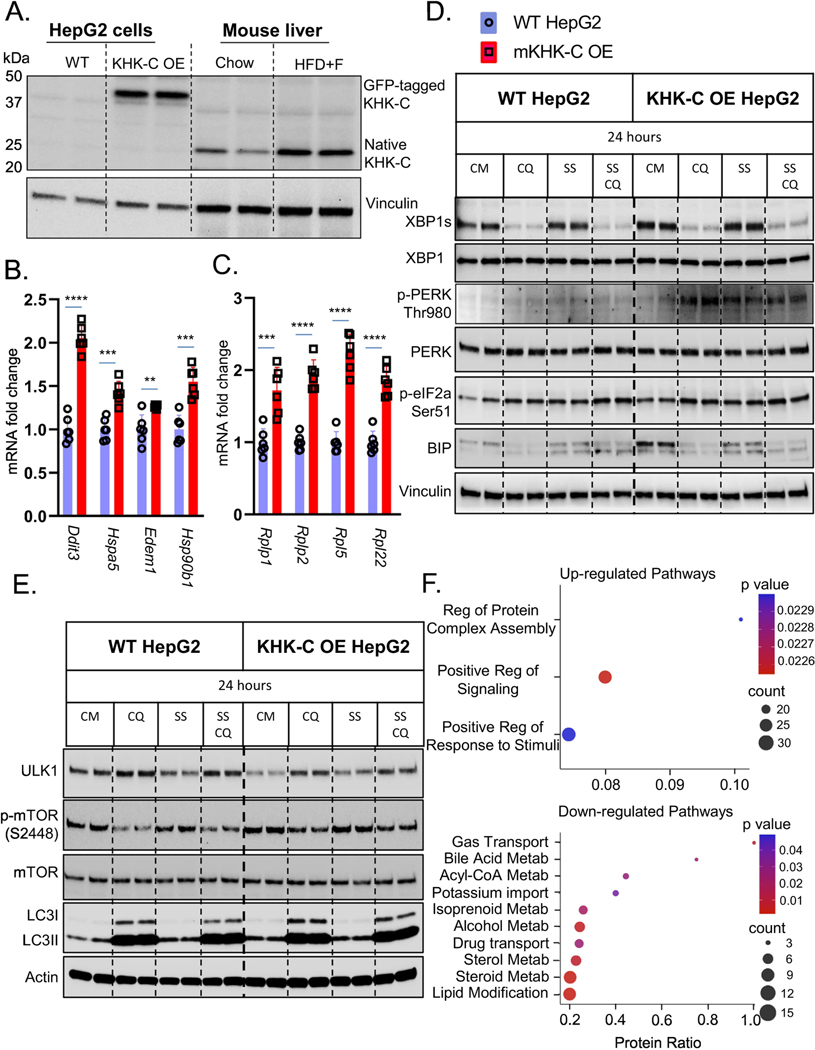
Figure 4. Overexpression of KHK-C Promotes ER Stress in vitro.
Wild type (WT) HepG2 hepatocytes were engineered to overexpress (OE) mouse KHK-C (mKHK-C OE) tagged with GFP. A) Western blot for KHK-C in WT and KHK-C OE cells as compared to liver homogenates from Chow and HFD+F-fed mice from Fig1. mRNA expresses as assessed by qPCR of B) ER-stress (Ddit3,Hspa5, Edem1,Hsp90b1), C) ribosomal (Rplp1,Rplp2,Rpl5,Rql22) genes were measured in WT HepG2 and mKHK-C OE cells, n=6 wells per group. D) WT HepG2 and mKHK-C OE hepatocytes were treated with complete media (CM), chloroquine (CQ), serum-starvation (SS), and a combination of serum starvation and chloroquine for 24 hours. Western blot was used to analyze D) ER-stress and E) autophagy pathways. mKHK-C was also OE in AML-12 mouse hepatocytes to minimize cell specific effects. Proteomic analysis using data-independent acquisitions on an Orbitrap Eclipse Tribrid platform was performed and most significantly F) upregulated and downregulated pathways were determined using ConsensusPathDB functional annotation pathway analysis. Statistics were completed by an unpaired Student’s t-test for qPCR data. **P≤0.01; ***P≤0.001; ****P≤0.0001. All data are presented as mean ± S.E.M.