Figure 1: DART-seq identifies SON as a m6A target in mouse HSCs. See also Figure S1 and Table S1–S3.
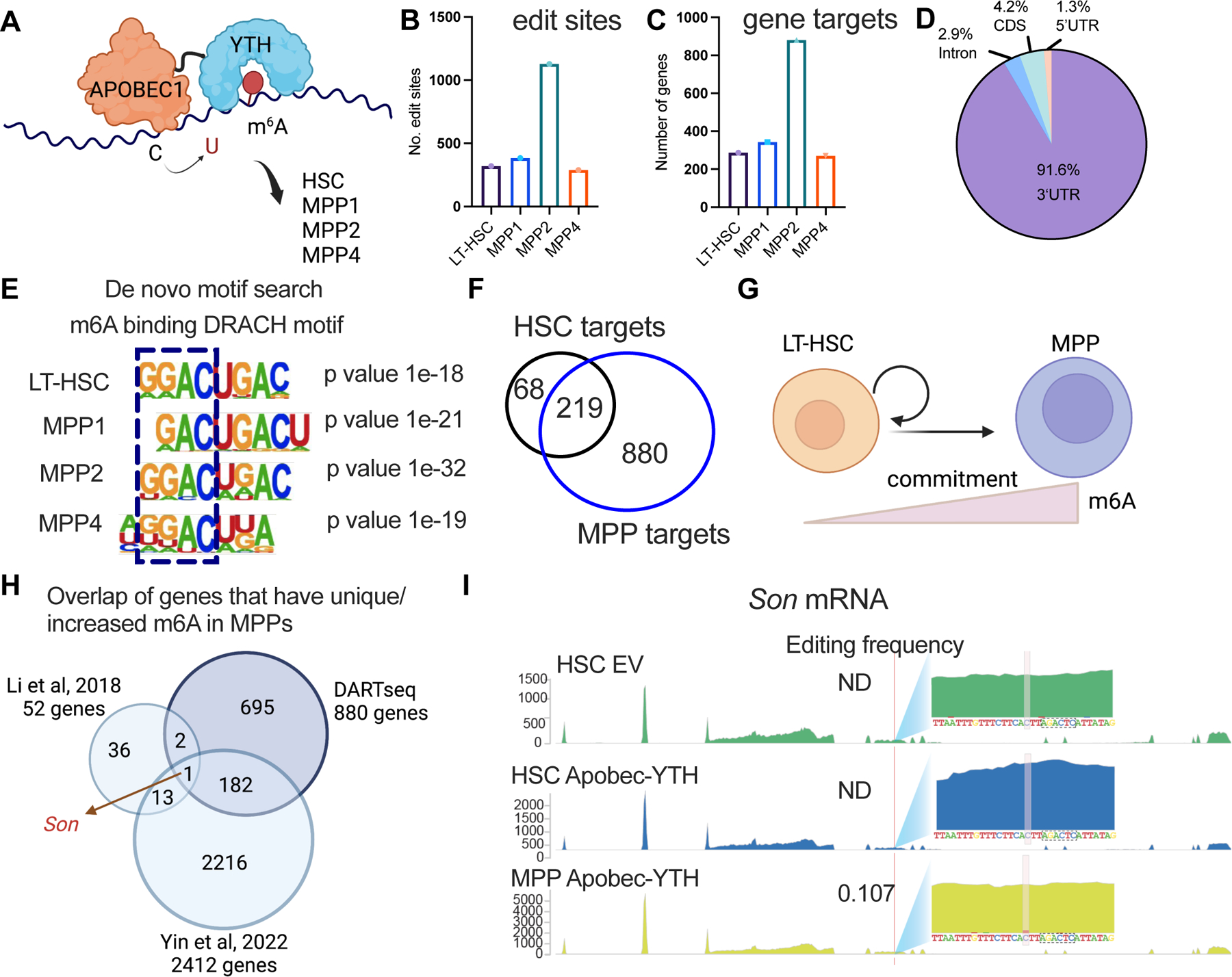
(A) DART-seq experimental scheme in sorted mouse HSC, MPP1, MPP2, and MPP4s.
(B) DART-seq identified significant edit sites from n=3 independent experiments. (sites with padj<0.1, differential editing > 0.1 were defined as significant sites).
(C) DART-seq identified significant target genes from n=3. independent experiments. (cut-off for significance: padj<0.1, differential editing > 0.1).
(D) DART-seq targets in mouse HSCs mainly localized to 3’UTR.
(E) De novo motif search analysis using edit sites in mouse HSCs and MPPs.
(F) Majority of HSC DART-seq targets overlap with MPP DART-seq targets.
(G) Scheme of genes whose m6A modifications increase from HSC to MPP commitment.
(H) Overlap of sites that have unique/increased m6A modification during HSC to MPP commitment with two other published datasets (m6A-seq31 and SLIMseq43).
(I) Representative track showing the C to U edit site on Son transcript and its adjacency to a DRACH motif. Editing frequencies in each sample was annotated. Yellow boxed ‘C’s were the edit sites and the sequences boxed with dotted black line were the adjacent DRACH motif. n=3 independent experiments. EV: empty vector.
