Fig. 5 |. Transcriptomic effects of oxycodone withdrawal are differentially expressed across reward-related brain regions with chronic SNI and Sham states.
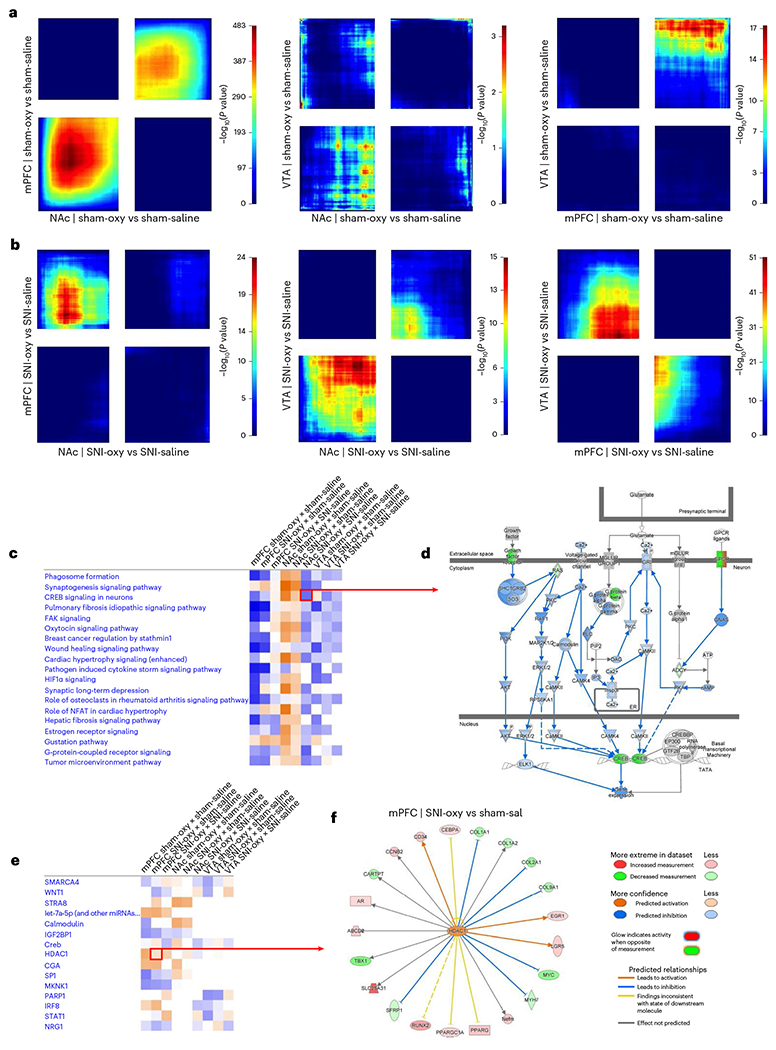
a,b, RRHO threshold-free comparisons of DEGs between the NAc, mPFC and VTA for oxycodone withdrawal with SNI and sham groups of adult male mice. Each pixel represents the overlap between the transcriptome of each comparison as noted, with the significance of overlap (−log10(P value)) of a hypergeometric test color coded. The lower-left quadrant includes co-upregulated genes, the upper-right quadrant includes co-downregulated genes, and the upper-left and lower-right quadrants include contra-regulated genes. Genes along each axis are sorted from most to least significantly regulated from the middle to outer corners. c, Top canonical pathways commonly regulated between Sham-Oxy versus Sham-Sal, SNI-Oxy versus Sham-Sal and SNI-Oxy versus SNI-Sal comparisons in the mPFC, NAc and VTA. d, Representation of significantly downregulated CREB signaling in neurons pathway in the NAc SNI-Oxy versus SNI-Sal condition. e, UR activity predictions across the aforementioned conditions/regions between oxycodone withdrawal under SNI and sham states. f, The genes predicted to be regulated by HDAC1 (UR) in the mPFC within the SNI-Oxy to Sham-Sal comparison.
