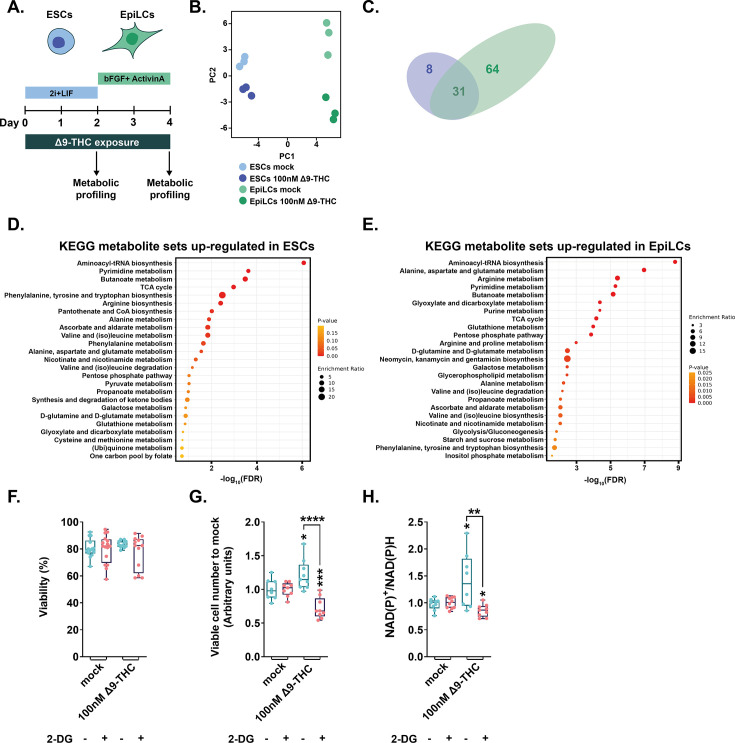
Figure 4. Δ9-THC-induced glycolysis sustain anabolism and ESCs proliferation (A) Diagram illustrating Δ9-THC exposure scheme and experimental strategy.
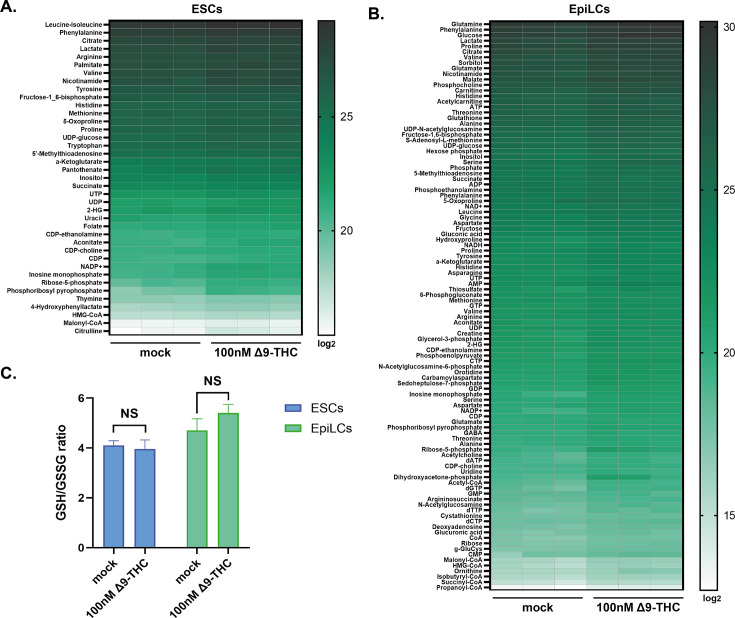
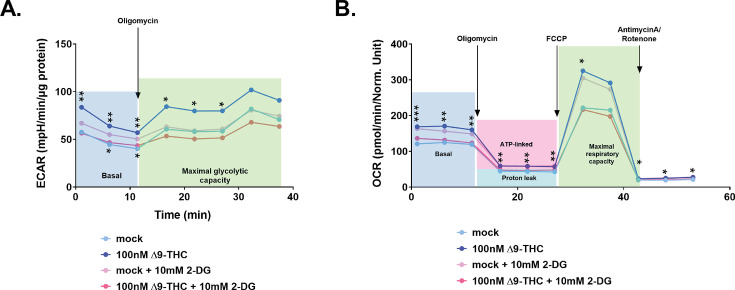
(B) PCA of the metabolomics profiling of either ESCs or EpiLCs mock-exposed or exposed to 100 nM Δ9-THC. (C) Venn diagram showing the overlap in upregulated metabolites following Δ9-THC exposure in ESCs and EpiLCs. (D and E) KEGG metabolite sets enrichment analysis for upregulated metabolites in ESCs and EpiLCs, respectively, performed by MetaboAnalyst (Pang et al., 2022). KEGG, Kyoto Encyclopedia of Genes and Genomes. (F) Whisker boxplot indicating the median cellular viability of stem cells exposed to 100 nM of Δ9-THC and 10 mM of 2-DG, as indicated, and their associated errors. (G) The median numbers of viable cells exposed to 100 nM of Δ9-THC and 10 mM of 2-DG, as indicated, were normalized to their own control (+/-2 DG). Median and associated errors were plotted in whisker boxplots. (H) The NAD(P)+/NADPH ratio of stem cells exposed to 100 nM of Δ9-THC and 10 mM of 2-DG, as indicated, was normalized to the one measured in the mock-treated condition (+/-2 DG). Median and associated errors were plotted in whisker boxplots. At least three independent biological repeats with three technical replicates (N=3, n=3). Statistical significance: *(p<0.05), **(p<0.01), ***(p<0.001), ****(p<0.0001).