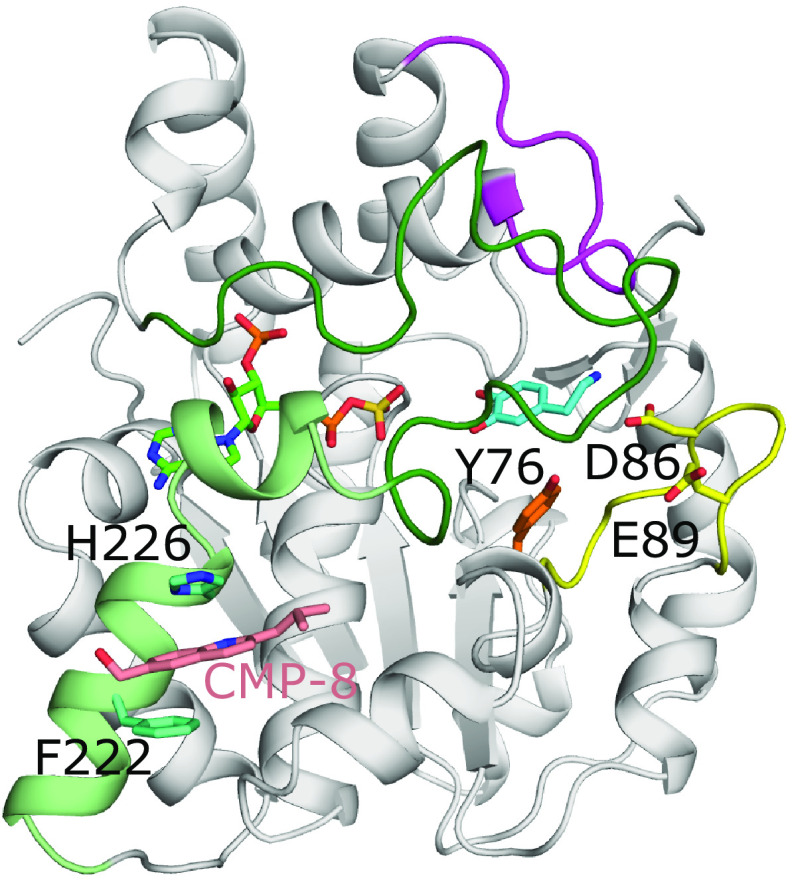
Fig. 8.
The allosteric binding sites of SULT1A3. Model of SULT1A3 with bound PAPS (green) and inhibitor CMP-8 (coral) bound to an allosteric site via aromatic stacking with His226 and Phe222 (model found at: https://www.modelarchive.org/doi/10.5452/ma-qtj80) (Darrah et al., 2019). The position of the acceptor substrate dopamine (cyan) was superimposed from PDB coordinates 2A3R (Lu et al., 2005). Also shown are residues Tyr76 (orange), Asp86, and Glu89 (both yellow) that compose the catechin-binding allosteric site that binds the monoamine neurotransmitter cofactor tetrahydrobiopterin (Cook et al., 2017). Loops 1, 2, and 3 are colored yellow, magenta, and dark green, respectively, as seen in SULT1E1 (Fig. 4). In the crystal structure of SULT1A3 without substrate present, residues 216-261 are disordered. This comprises loop 3 and the two helices (light green) N-terminal to loop 3 that make up one of the allosteric binding sites (PDB code 1CJM) (Bidwell et al., 1999).