Figure 7.
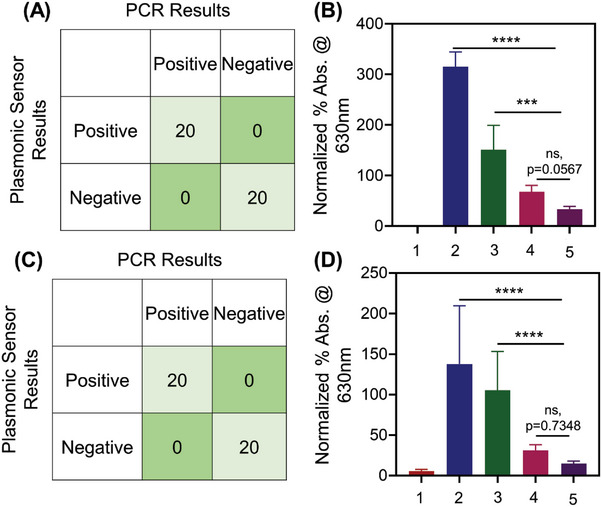
Chlamydia and Gonorrhea detection from forty (n = 40) deidentified clinical samples using CHAI buffer without any added purification steps. (A) Confusion matrix obtained from the studied ssDNA‐AuNPs targeting CT DNA. Results from forty (n = 40) deidentified clinical samples were validated with the gold standard technique qPCR. (B) Graphical representation of normalized percentage (%) absorbance change at 630 nm with CT targeted ssDNA‐AuNPs for 1. Control (water sample); 2. CT positive sample (n = 10); 3. both CT positive and NG positive sample (n = 10); 4. NG positive sample (n = 10) and 5. both CT and NG negative sample (n = 10). Data are presented as mean ± SD. The one‐way ANOVA showed that the assay response was significantly different between positives and negatives. ****p‐value <0.0001, ***p = 0.001, ns; p = 0.0567. (C) Confusion matrix obtained from the studied ssDNA‐AuNPs targeting NG DNA. Results from forty (n = 40) deidentified clinical samples were validated with the gold standard qPCR. (D) Graphical representation of normalized percentage (%) absorbance change at 630 nm with NG targeted ssDNA‐AuNPs for 1. Control (water sample), 2. NG positive sample (n = 10), 3. both CT positive and NG positive sample (n = 10), 4. CT positive sample (n = 10) and 5. both CT and NG negative sample (n = 10). Data are presented as mean ± SD. The one‐way ANOVA showed that the assay response was significantly different between positives and negatives. ****p‐value <0.0001, ns; p = 0.7348.
