Figure 5.
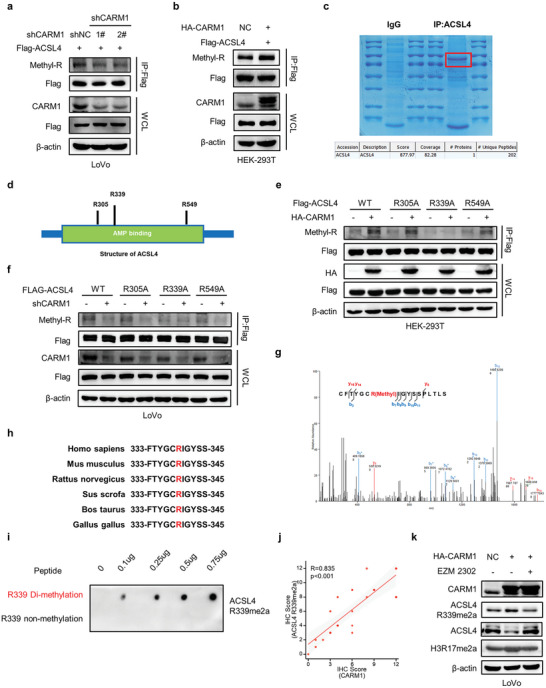
CARM1 methylates ACSL4 at R339. a,b) Co‐immunoprecipitation (Co‐IP) was performed to detect the methylation levels of ACSL4 with CARM1 attenuation (left) or upregulation (right). c) IP assay was performed for the enrichment of ACSL4 protein, staining was performed with Coomassie bright blue and verification was conducted by using mass spectrometry. d) Schematic diagram of ACSL4 structure and methylation sites. e) Co‐IP was performed to detect the methylation changes in WT ACSL4 and the R305A, R339A, and R549A mutants with CARM1 overexpression. f) Co‐IP was performed to detect the methylation changes in WT ACSL4 and the R305A, R339A, and R549A mutants with CARM1 attenuation. g) Secondary mass spectrometry result of one possible methylation residue at arginine 339. h) The ACSL4 R339 site amino acid in different species. i) Dot plot assay verifying the specificity of anti‐ACSL4 R339me2a using 0.1‐0.75 µg of different peptides. j) Correlation between CARM1 and ACSL4 R339Ame2a expression in colorectal cancer (CRC) tissues (n = 25) was determined by using the Spearman correlation coefficient test. All p and R values were calculated with Spearman's r test. k) Western blot analysis of vector‐ and CARM1‐overexpressing LoVo cells treated with DMSO or 10 × 10−9 m EZM2302 for 24 h. Protein levels of CARM1, ACSL4, ACSL4 R339me2a, and H3R17me2a were assayed.
