Figure 6.
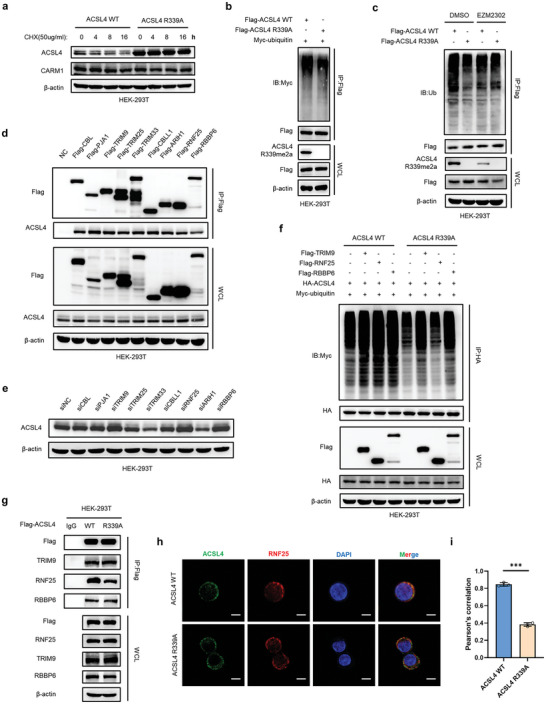
ACSL4 R339 methylation promotes the degradation of ACSL4 by RNF25. a) Western blot analysis of ACSL4 WT and ACSL4 R339A‐overexpressing stable HEK293T cells treated with 50 µg mL−1 cycloheximide for the indicated times. b) HEK293T cells transfected with the indicated plasmids and treated with 10 × 10−6 m MG132 for 8 h. Immunoprecipitation (IP) with anti‐Flag antibody and Western blotting with anti‐Myc antibody were performed to detect the ubiquitination level of ACSL4. c) HEK293T cells were treated as in (b) and treated with or without 10 × 10−9 m EZM2302 for 24 h. IP with an anti‐Flag antibody and Western blotting with an anti‐ubiquitin antibody were performed to detect the ubiquitination level of ACSL4. d) IP analyses were performed to examine the interaction between the indicated E3 ubiquitin ligase and endogenous ACSL4 using anti‐Flag antibodies in HEK293T cells. e) Western blot analysis of HEK293T cells transfected with the indicated siRNAs of E3 ubiquitin ligase. Protein levels of ACSL4 were assayed. f) HEK293T cells transfected with the indicated plasmids and treated with 10 × 10−6 m MG132 for 8 h. IP with anti‐HA antibody and Western blotting with anti‐Myc antibody were performed to detect the ubiquitination level of ACSL4. g) IP analyses were performed to detect the interaction changes between ACSL4 WT or ACSL4 R339A and the indicated E3 ubiquitin ligase. h) Immunofluorescence staining was performed to observe the colocalization changes of ACSL4 (green) and RNF25 (red) in ACSL4 WT and ACSL4 R339A LoVo cells. The nucleus is labeled by using DAPI (blue). Scale bar, 20 µm. i) Statistics of the colocalization of ACSL4 and RNF25, as indicated by Pearson's correlation (30 cells per sample). The data shown represent the mean ± SD. Comparisons were made by using two‐tailed, unpaired Student's t‐test; *** p < 0.001.
