Figure 1.
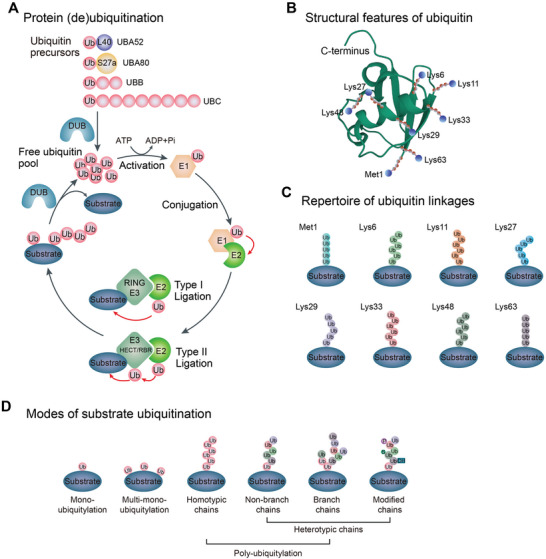
The ubiquitination cascade and the complexity of ubiquitin modifications. A) Schematic showing the key events in ubiquitination and deubiquitination. Ubiquitin (Ub) is produced by cleavage of Ub precursors via deubiquitylating enzymes (DUBs). Ubiquitin precursors include four different genes: UBB and UBC genes code for a polyubiquitin precursor and UBA52 and UBA80 genes code for a single copy of Ub fused to the ribosomal proteins L40 (L) and S27a (S), respectively. Ubiquitin is activated by the E1‐activating enzyme in an ATP‐dependent manner. The E2‐conjugating enzyme catalyzes the conjugation of E1‐bound Ub to itself. Finally, E3 ligase directly (RING E3 ligases) or indirectly (HECT/RBR E3 ligases) transfers E2‐bound Ub to a substrate, forming a thioester linkage between the carboxyl group of glycine in Ub and the ε‐amino group of lysine in the substrate. Ubiquitin molecules are removed from substrates by DUBs and recycled. B) Schematic representation of the structure of Ub. Stick representation showing the seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) and Met1 of Ub (Protein Data Bank ID: 1UBQ[ 612 ]), and amino groups that are modified with Ub during the formation of the chain are marked with blue spheres. C) The repertoire of ubiquitin linkages, Met1, and seven Lys residues in Ub can form specific chain linkages with distinct conformations and exert specific (or unknown) functions. D) A substrate can be modified by mono‐, multi‐mono‐ or poly‐ubiquitin. Ubiquitin chains are colored according to linkage type, and each type of chain represents a distinct posttranslational modification. Polyubiquitin includes homotypic chains and heterotypic chains. Heterotypic chains are either branched or modified, such as SUMOylation (S), phosphorylation (P), or hydroxylation (OH), etc.
