Figure 4.
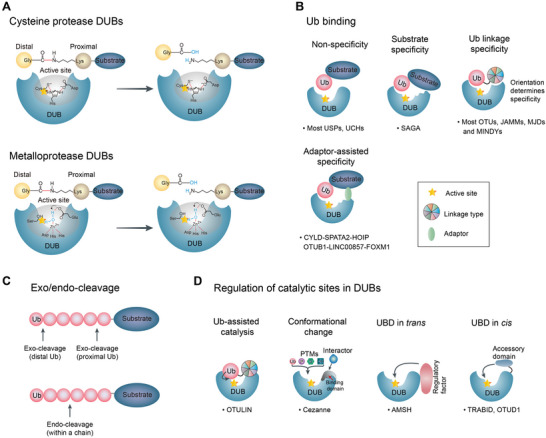
Catalytic DUB mechanisms of cleavage and recognition. A) Cysteine protease DUBs commonly include a catalytic pocket composed of cysteine, histidine, and an acidic residue. JAMM family DUBs are Zinc (Zn)‐dependent metalloproteases. A Zn atom is essential for the active site that is coordinated by conserved catalytic residues and a water molecule. Red represents a fragile isopeptide. B) Principles of substrate recognition and linkage specificity according to a variety of Ub binding and regulation catalytic sites of DUBs. Most members of the USP and UCL subfamilies have nonspecific interactions with the substrate (without a motif). Conversely, SAGA has substrate specificity through motif interactions with the substrate protein. Ub linkage specificity is acquired so that only one linkage type is identified by Ub‐binding sites to catalyze DUB activity. Many members of OTU‐, JAMM‐, Josephin‐, and MINDY families have linkage specificity. In addition, some DUBs recognize targeted proteins with the assistance of adaptors or scaffolds. C) The characteristics of Ub‐binding sites in DUBs determine whether polyubiquitin is cleaved from the distal‐ or the proximal end (exo‐cleavage) or within the en bloc chain (endo‐cleavage). D) Regulation of catalytic sites in DUBs. Ub complements the enzymatic activity by directly participating in the active site, and other post‐translational modifications (SUMOylation, phosphorylation, and hydroxylation) are also key features in regulating DUB activity. Furthermore, regulatory factor in trans (between two molecules) or accessory domain in cis (within a molecule) exerts a boosting effect on the catalytic activity. The star indicates the active site.
