Extended Data Fig. 6. Structure and functions of DNA and RNA aptamers.
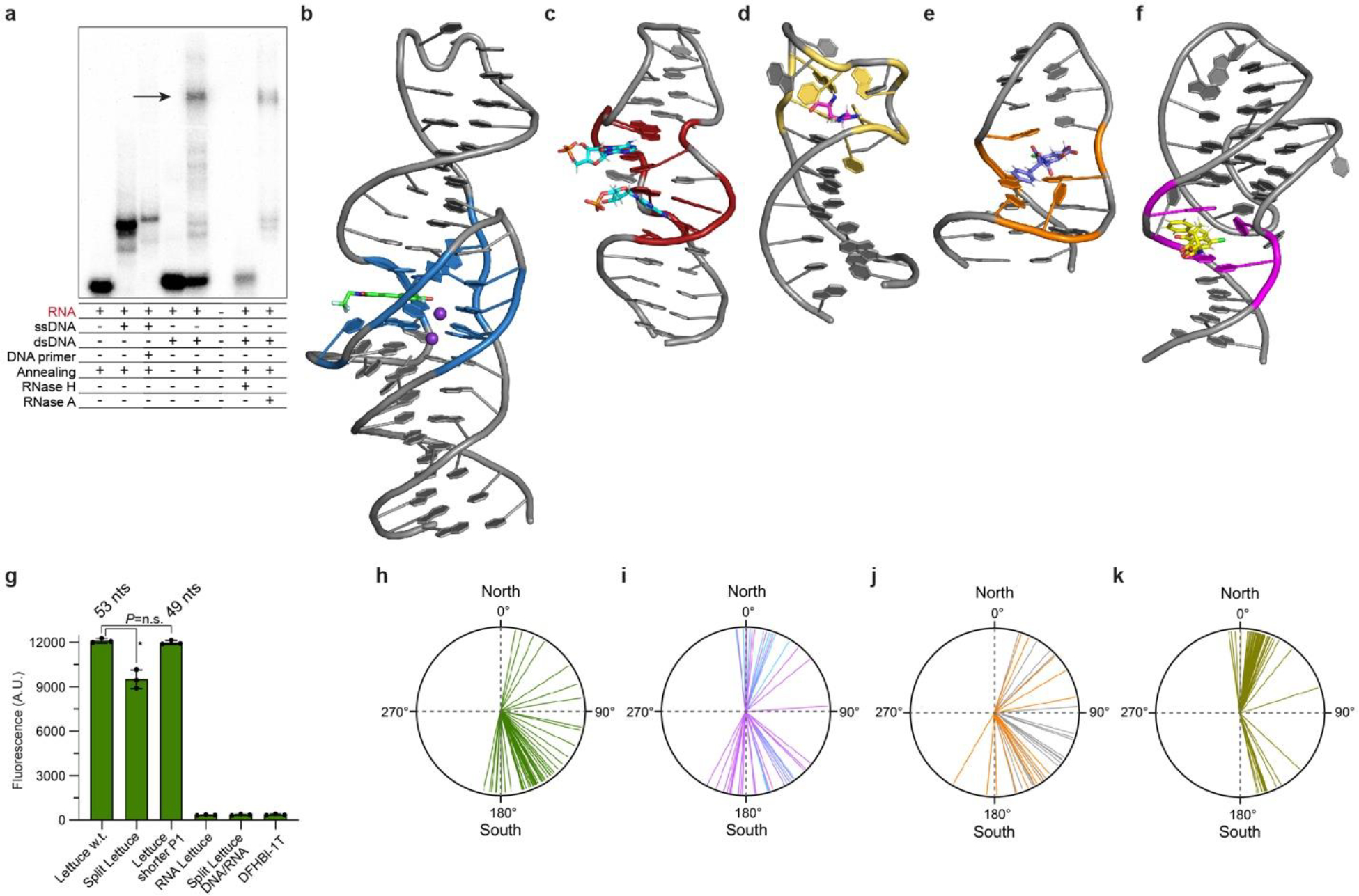
a, Autoradiogram of 6% native polyacrylamide gel electrophoresis demonstrating R-loop formation (arrow). b, Lettuce–DFHBI-1T co-crystal structure (this study). c, NMR structure of the AMP-binding DNA aptamer in complex with two AMP molecules [1; PDB:1AW4]. d, NMR structure of a DNA aptamer bound to argininamide [2; PDB:1DB6]. e, NMR structure of the OBA3 DNA aptamer bound to ochratoxin A [3; PDB:6J2W]. E, NMR structure of the OBA36 DNA aptamer bound to ochratoxin A [4; PDB:7W9N]. f, Fluorescence of wild=type Lettuce w.t. used in the crystallographic studies, Split Lettuce, Lettuce with shorter P1 used in fluorescence studies (Extended Data Table S4), RNA comprised of the Lettuce sequence, and split Lettuce comprised by DNA (34 nts) and RNA (16 nts of 3’ terminus) in the presence of DFHBI-1T (mean ± s.d., n = 3). Asterisk denotes P= 0.002 (two-sided t-test). No significant difference (P= 0.407) between Lettuce w.t. and Lettuce with shorter P1 (two-sided t-test). g, Pucker angles of each deoxynucleotide of Lettuce–fluorophore complexes (mean of 8 structures). h, Pucker angles of each deoxynucleotide of the RNA-ligating deoxyribozyme 9DB1 [5; PDB:5CKK]. Puckers of core deoxynucleotides are in magenta; puckers of deoxynucleotides that hybridize to the RNA product strand are in cyan. j, Pucker angles of each deoxynucleotide of the RNA-cleaving deoxyribozyme Dz36 [6; PDB: 5XM8]. Puckers of core deoxynucleotides are in orange; puckers of deoxynucleotides that hybridize to the RNA substrate strand are in gray. k, Pucker angles for each ribose of the RNA fluorogenic aptamer iSpinach in complex with DFHBI [7; PDB:5OB3]. Individual puckers for all four aptamers are in Extended Data Table 2. Puckers calculated using ref 8.
