Figure 2.
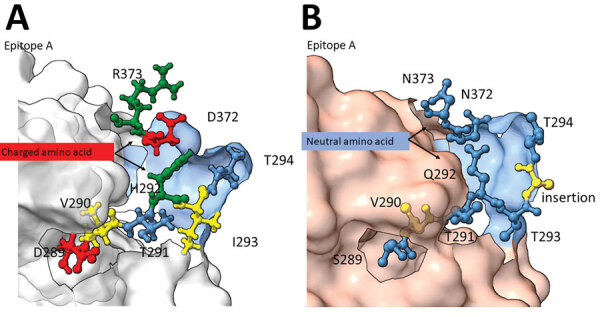
Structural changes of emergent novel norovirus GII.4 strains from 3 continents. A) Sydney GII.4 strain (GenBank accession no. JX459908); B) GII.4 San Francisco strain. The 3-dimensional structure models were predicted by using ChimeraX version 1.4 (11) and the alphafold prediction tool (12). Models show structural changes near and within the epitope A antigenic region on GII.4 San Francisco P-domain (panel B) are overlayed on a GII.4 Sydney 2012 backbone (Protein Data Bank, https://www.rcsb.org/structure/4OP7). Negatively (red) and positively (green) charged amino acids of GII.4 Sydney (panel A) were replaced with neutral amino acids (blue) in the GII.4 San Francisco strain and a hydrophobic (yellow) amino acid, alanine, was inserted between T293 and T294.
