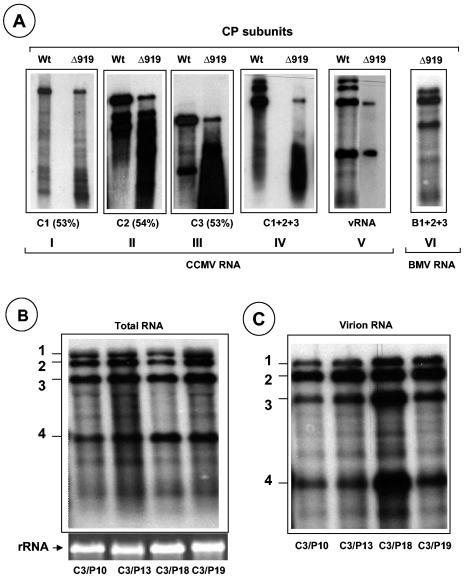
FIG. 5.
Encapsidation competence of CCMV RNAs with N-terminal ARM mutants. (A) In vitro assembly assays. Northern blot analysis of RNA isolated from virions assembled in vitro with Δ919 CP subunits and each of the three genomic RNA transcripts (panels I to III), their mixture (panel IV), virion RNA (panel V), and a mixture containing all three BMV RNA transcripts (panel VI). Purified CP subunits and the indicated RNAs were allowed to assemble in vitro as described in Materials and Methods. Conditions for denaturizing RNA, electrophoresis, and hybridization with riboprobes are as described in the legend of Fig. 3. RNA samples shown in panels I to V were hybridized with a probe complementary to the 3′ end of CCMV RNA, whereas the RNA sample shown in panel VI was hybridized with a probe complementary to the 3′ end of BMV RNA. The numbers shown in parentheses below panels I to III represent the percentages of assembly efficiency of Δ919 CP subunits for each WT RNA transcript with respect to WT CP subunits. (B and C) Packaging profiles of CCMV CP bearing N-terminal proline mutations (Fig. 1A). Shown are Northern blots of total nucleic acids or virion RNA recovered from symptomatic leaves of cowpea infected with the indicated C3 mutant. The blots were hybridized with 32P-labeled RNA probes complementary to the homologous 3′ region present on each of the four CCMV RNAs. The positions of CCMV RNAs are shown to the left of each blot.