Abstract
Proline-rich antimicrobial peptide (PrAMP) Drosocin (Dro) from fruit flies shows sequence similarity to other PrAMPs that bind to the ribosome and inhibit protein synthesis by varying mechanisms. The target and mechanism of action of Dro, however, remains unknown. Here we show that Dro arrests ribosomes at stop codons, likely sequestering class 1 release factors associated with the ribosome. This mode action is comparable to that of apidaecin (Api) from honeybees, making Dro the second member of the Type II PrAMP class. Nonetheless, analysis of a comprehensive library of endogenously expressed Dro mutants shows that the interactions of Dro and Api with the target are markedly distinct. While only few C-terminal amino acids of Api are critical for binding, the interaction of Dro with the ribosome relies on multiple amino acid residues distributed throughout the PrAMP. Single residue substitutions can significantly enhance the on-target activity of Dro.
Keywords: ribosome, antibiotics, proline-rich antimicrobial peptides, apidaecin, release factor
Introduction
Antimicrobial peptides (AMPs) are components of the innate immune system of higher organisms helping them to suppress infections caused by bacterial pathogens 1. The best-studied AMPs kill bacteria by attaching to the bacterial cytoplasmic membrane, disrupting its integrity and causing cell lysis 1. However, some AMPs penetrate the bacterial cell and inhibit its growth by acting upon intracellular targets 2. Proline-rich AMPs (PrAMPs) belong to the latter class 3,4.
The known PrAMPs, produced by insects and mammals, range in size from ~15 to ~60 amino acids 4,5. However, their pharmacophore (the sequence critical for the on-target activity) is usually limited to a smaller segment, in some cases, of only 12 amino acids long 6,7. Most of the studied PrAMPs are produced as unmodified peptides, but some carry posttranslational modifications 8,9. Specific peptide transporters import PrAMPs into the cytoplasm of the bacterial cell 10,11.
While it was initially proposed that PrAMPs stop bacterial growth by inhibiting the activity of the chaperone DnaK 12, subsequent findings demonstrated that DnaK-lacking mutants remain sensitive to PrAMPs 13–15. More recent biochemical and structural studies showed that PrAMPs bind to the ribosome and inhibit its functions 15–21. The finding that ribosomal mutations confer resistance to some PrAMPs 7,19,21, firmly established the ribosome as the primary target of PrAMPs in bacteria.
PrAMPs bind in the nascent peptide exit tunnel (NPET) in the large ribosomal subunit 17–21, which serves as the passageway for the growing polypeptide assembled in the peptidyl transferase center (PTC). Besides catalyzing peptide bond formation, the PTC is also responsible for the release of the completed protein from the P-site tRNA in the reaction involving class-1 release factors (RF1 and RF2 in bacteria). According to their mechanism of action, the examined PrAMPs fall into two major categories 4. Many of them, including oncocin, bactenecin, pyrrhocoricin and metalnikowin, belong to Type I. Their N-termini invade the PTC A site while the C-termini protrude down the NPET 17–20 (Fig. 1a). Ribosomes associated with Type I PrAMPs can form initiation complexes but cannot accommodate an elongator aminoacyl-tRNA and catalyze formation of the first peptide bond. As a result, Type I PrAMPs arrest ribosomes at start codons 17–20,22,23. The only known PrAMP belonging to Type II, apidaecin (Api), acts in a distinctly different way. The synthetic Api derivative, Api137 24, also binds in the NPET, but in contrast to Type I PrAMPs, in an orientation that mimics that of a nascent polypeptide, i.e., with the N-terminus stretched down the tunnel and the C-terminus bordering the PTC 21,25 (Fig. 1a). Api acts upon the ribosome that has reached the stop codon and has released the completed protein but is still associated with class 1 RFs 21,26. Once the newly made polypeptide vacates the ribosome, Api penetrates the NPET where its C-terminal residue interacts with the 3’ ribose of the P-site-bound tRNA while the penultimate arginine residue (R17) engages with the RF still bound to the PTC A site 21. As a result, Api freezes the ribosome at the stop codon and sequesters class 1 RFs 21,25. Due to the excess of ribosomes over RFs, the pool of free RFs is rapidly depleted and, therefore, the rest of the ribosomes are unable to release the completed proteins when they reach the stop codons. This leads to ribosome stalling in a pre-release state and eventual stop codon bypass by aminoacyl-tRNA misincorporation or frameshifting 7,21,26.
Fig. 1: The PrAMP Drosocin stalls ribosomes at stop codons during in vitro translation.
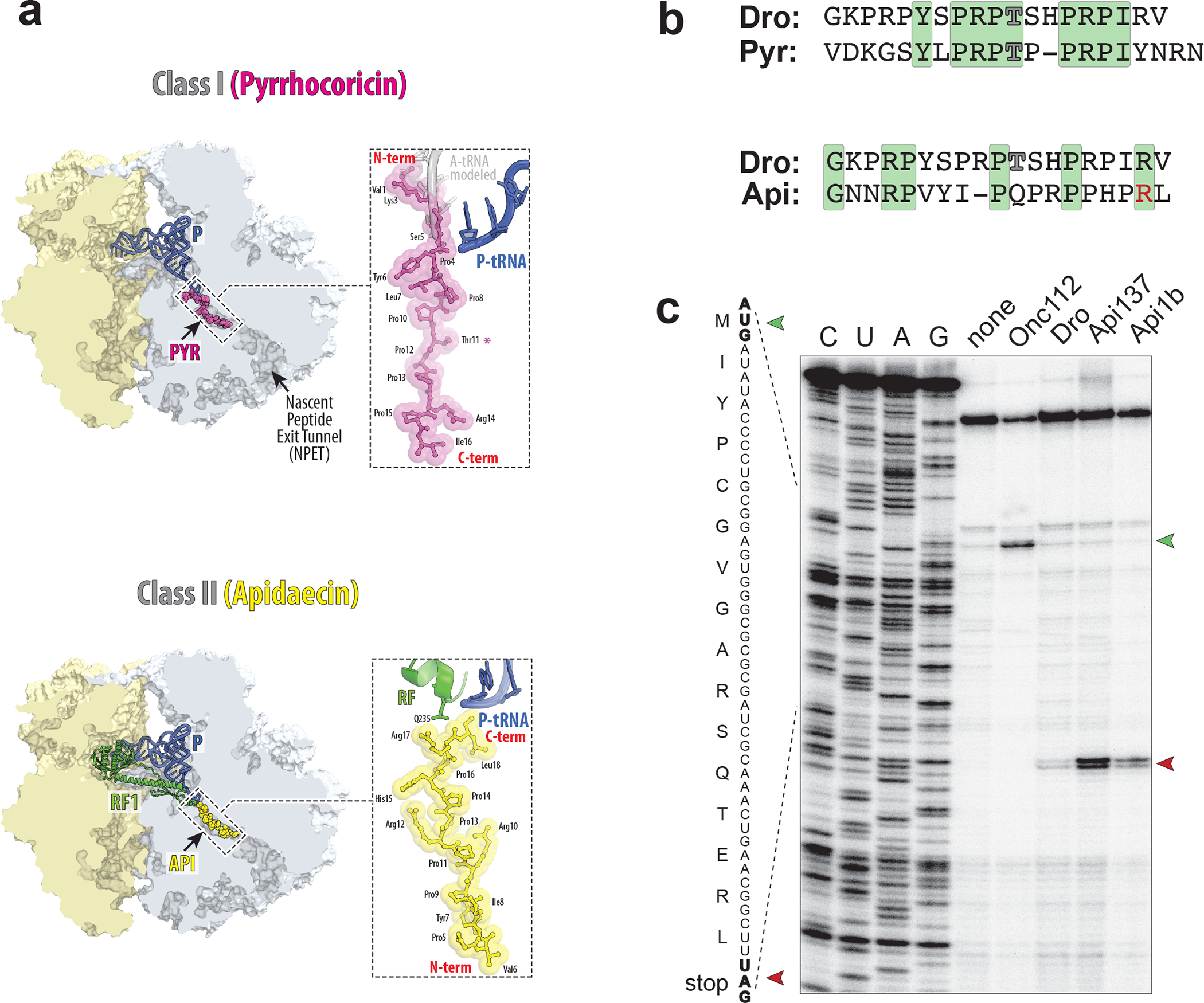
a, Comparison of binding of Type I PrAMP pyrrhocoricin (Pyr) (PDB 5HD1 19) and Type II PrAMP apidaecin (Api) (PDB 5O2R 21) to the bacterial ribosome. b, The Drosocin (Dro) amino acid sequence, aligned with that of Type I PrAMP Pyrrhocoricin (Pyr) or Type II PrAMP Apidaecin (Api). Residues identical between the compared PrAMPs are highlighted in green. The glycosylated Thr (T) residues of Dro and Pyr are indicated by outlined characters. The functionally critical penultimate Arg (R) residue of Api is highlighted in red. c, Toeprinting analysis of the ribosome arrest at the UAG stop codon of a model ORF due to the addition of synthetic PrAMPs Dro, Api137, or Api1b. Type I PrAMP Onc112 that arrests ribosomes at start codons 19, was used as a control. The reaction where no PrAMPs were added is labeled as “none”. The toeprint bands of ribosomes arrested at the start or stop codons of the ORF are marked with green or red arrowheads, respectively. Sequencing reactions are labeled as C, U, A, G. Shown is a representative gel of three independent experiments that produced converging results.
No other PrAMP is known to act in an Api-like fashion. However, it was noted that drosocin (Dro), a PrAMP encoded in the genome of the fruit fly Drosophila melanogaster and exhibiting promising activity against a range of Gram-negative and some Gram-positive bacteria 16,27–32, shows some sequence similarity to Api and may compete with Api for binding to the ribosome 16. In the 19-amino acid long sequence of Dro, GKPRPYSPRPTSHPRPIRV, Thr11 (underlined) is O-glycosylated with N-acetylgalactosamine and galactose 8 but Dro variants lacking this modification retain antibacterial activity with a slight (up to 8-fold) increase in minimal inhibitory concentration (MIC) 8,27,29.
While Dro is one of the first discovered PrAMPs, little is known about its mode of action. Although Dro binds to the ribosome with a moderate affinity 16, it remains unclear whether it inhibits cell growth by interfering with translation. Furthermore, even if the ribosome is the primary target of Dro, it is unclear which step of translation might be affected and through which mechanism.
Here we unraveled the mechanism of action of Dro in the bacterial cell and identified the amino acid residues critical for target engagement and activity. We show that the unmodified Dro interferes with translation termination and retains its inhibitory activity when expressed directly in bacterial cells. By directly expressing Dro in bacteria, we surveyed all possible single amino acid substitutions and identified the residues critical for the PrAMP’s ribosome-targeting activity. Furthermore, we found mutants whose activity is superior of that of the unmodified wild-type (wt) Dro. Our findings establish that peptides with diverse sequences can interfere with the termination stage of translation by binding in the ribosomal NPET.
Results
Dro arrests the ribosome at the stop codon of the ORF
Native Dro shows moderate sequence similarity with the Type I PrAMP pyrrhocoricin, which also carries a glycosylated threonine 9 (Fig. 1b). This may suggest that like Type I PrAMPs, Dro could invade the PTC A site with its N-terminal segment and inhibit formation of the first peptide bond 17,19 (Fig. 1a). Sequence resemblance between Dro and the Type II PrAMP Api (Fig. 1b) is less pronounced, but the presence of a penultimate arginine residue in both PrAMPs, which plays a pivotal role in the activity of Api 7,21, leaves the possibility that Dro’s mode of binding and inhibition could be similar to that of Api 16 (Fig. 1a). To establish the genuine mechanism of Dro action, we tested its effects on protein synthesis in a cell-free translation system. Because glycosylation of the Thr11 residue is neither strictly required for the antimicrobial activity of Dro 8,27,29 nor is critical for the interaction of the PrAMP pyrrhocoricin with the ribosome 19, we avoided the rather cumbersome synthesis of the glycosylated peptide and carried out all our experiments with the unmodified synthetic Dro.
In agreement with published results 16,33, Type I PrAMP Onc112, which interferes with translation initiation 17,19, efficiently inhibits in vitro expression of GFP rather independently of the presence of RFs in the reaction (Extended Data Fig. 1a). Similar to the termination inhibitor Type II PrAMP Api137, Dro very mildly inhibits the GFP production in reactions depleted of RFs (under these conditions, GFP expression possibly relies on stop codon readthrough) (Extended Data Fig. 1a, light gray bars). In contrast, and also comparable to the effect observed with Api137, the inhibitory activity of Dro becomes more evident when the reactions are supplemented with RFs (Extended Data Fig. 1a, dark gray bars). Notice however, that even in the presence of RFs, Dro inhibits GFP expression rather modestly, achieving ~60% inhibition at the highest concentration tested (50 μM) (Extended Data Fig. 1a). Thus, these data suggest that, similar to Api137, Dro might act upon translation termination since its inhibitory action requires the presence of class 1 RFs.
To further explore the possibility that Dro interferes with translation termination, we tested its effects on the progression of ribosomes through mRNA by toeprinting, a technique that can map sites of inhibitor-induced ribosome stalling during in vitro translation 34. Consistent with previous reports, Type I PrAMP Onc112 stalls ribosomes at the start codon of the open reading frame (ORF) 17,19, whereas Api137 or its native prototype Api1b, arrests translation at the stop codon 21 (Fig. 1c). Addition of Dro had no effect on translation initiation, but a discernible toeprint band corresponding to ribosomes arrested at the UAG stop codon appeared (Fig. 1c). Thus, toeprinting analysis provided strong evidence that Dro inhibits termination of translation. When we modified the template used in the toeprinting reaction by replacing the UAG stop codon, recognized by RF1, with UGA, recognized by RF2, the intensity of the toeprint band representing Api- or Dro-induced arrest of the terminating ribosome significantly diminished (Extended Data Fig. 1b). This was not surprising because RF2 present in the toeprinting reaction carries the E. coli K12 strain-specific polymorphism resulting in replacement of Ala246 with Thr 35, a substitution known to render RF2 more resistant to the action of Api 21. Apparently, the Ala246Thr substitution also diminishes sensitivity of RF2 to Dro.
Dro stops cell growth by inhibiting translation termination
Our in vitro experiments show that Dro stalls ribosomes at stop codons (Fig. 1; Extended Data Fig. 1) but do not prove that inhibition of translation termination is the key mode of Dro action against bacterial cells, especially considering that Dro had been previously shown to interfere with the activity of the DnaK chaperone 12,36. To identify the primary target of Dro action in bacteria, we tested its effect upon several previously characterized mutants. We first compared Dro’s MIC against the E. coli K12 strain BW25113 and its ΔdnaK derivative. Although the parental strain is fairly tolerant to Dro due in part to the intrinsic Ala246Thr mutation in the RF2 gene 35, it can be readily inhibited with 32–64 μg/mL of Dro. Deletion of the dnaK gene did not alter the Dro MIC, thereby excluding DnaK as the main target. We further determined Dro’s MIC against the mutants of the E. coli B strain BL21 carrying alterations in ribosomal protein uL16 (the R18C substitution) or in RF2 (the R262C or Q280L mutations), isolated previously for their resistance to Api 21. (Of note, RF2 of the wt BL21 cells has an ‘unmutated’ Ala246 conserved in other enterobacteria). All the three tested mutants showed significantly increased resistance to Dro compared to the wt strain (Table 1). These results strongly suggest that the ribosome is the primary target of Dro in bacterial cells and that this PrAMP inhibits bacterial growth by functionally interacting with ribosomes and RFs.
Table 1.
Antibacterial action of Dro involves the ribosome and RFs.
Dro induces stop codon readthrough by sequestering RFs
Api-mediated sequestration of RFs on a small fraction of cellular ribosomes prevents the rest of the ribosomes from releasing the nascent proteins at stop codons, leading to the eventual stop codon bypass by amino acid misincorporation 21,26. To test whether Dro action in bacteria leads to stop codon readthrough, we used E. coli BL21 cells transformed with a reporter plasmid, in which in-frame fused rfp and gfp genes are separated by the UGA stop codon 37. Placing drops of PrAMPs solutions on agar plates inoculated with a lawn of cells generated a gradient of the inhibitor concentration producing a clearing zone of no cell growth. At subinhibitory concentrations, both Dro and Api, albeit not the control antibiotic chloramphenicol (Chl), readily induced GFP expression while synthesis of the reference RFP protein remained unaltered (Fig. 2a). Similar results were obtained in liquid culture experiments where cells carrying the reporter were exposed to varying sub-MIC concentrations of Dro and Api (Fig. 2b). We concluded that both PrAMPs greatly stimulate stop codon readthrough, thereby revealing that, similar to Api, Dro likely sequesters RFs on terminating ribosomes.
Fig.2: Dro induces stop codon readthrough.
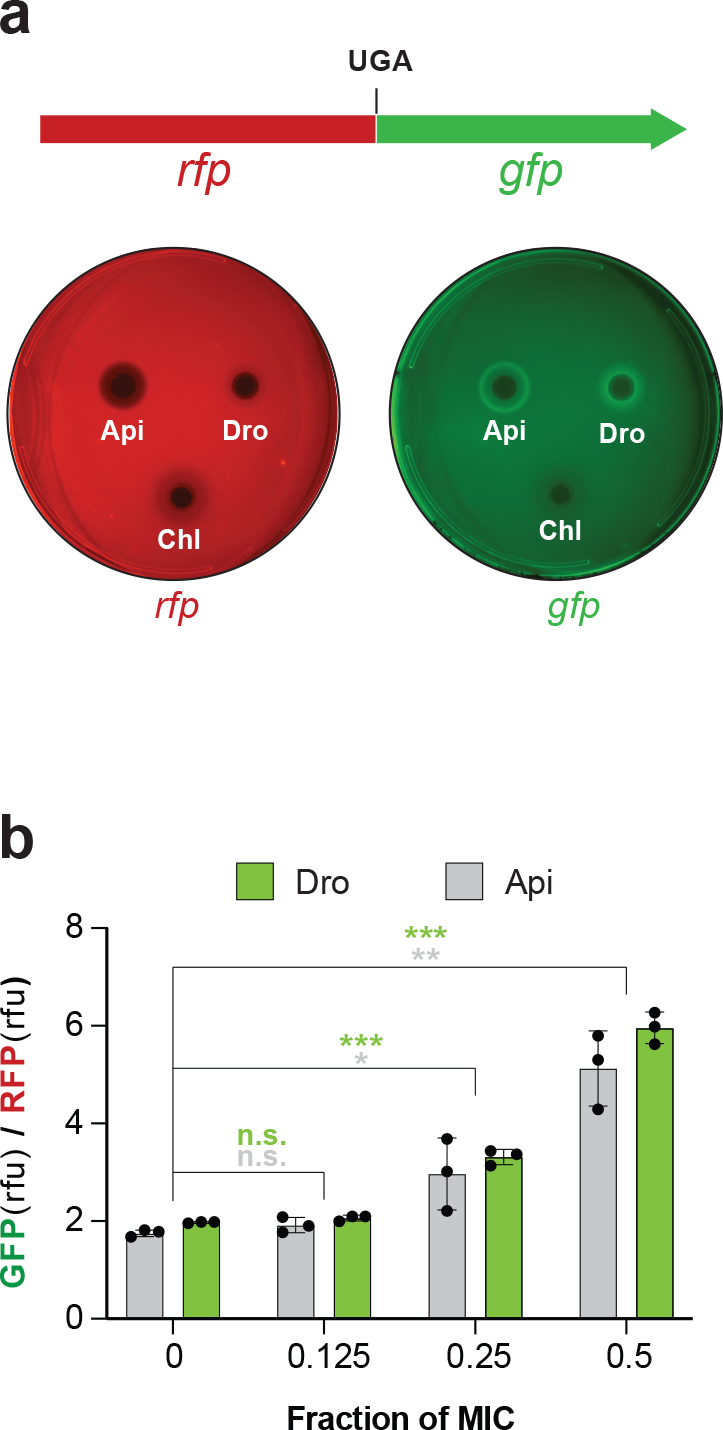
a, Top: Schematics of the rfp-gfp reporter in the pRXG plasmid 37 where expression of GFP requires readthrough of the stop codon separating the two genes. Bottom: Visualization of the Dro-mediated readthrough activity. Drops containing Dro (9 μg), the control readthrough inducing PrAMP Api137 (1 μg), or the non-inducing antibiotic chloramphenicol (Chl) (1 μg) were placed onto agar plates inoculated with a lawn of E. coli BL21 cells carrying the pRXG[UGA] reporter plasmid. The brighter GFP fluorescence halos on the right plate imaged in the GFP (Cy2) channel indicate Dro- or Api137-induced stop codon readthrough in cells growing at subinhibitory concentrations of the PrAMPs. The lack of similar halos on the plate imaged in the RFP (Cy3) channel demonstrates even expression of the reference RFP protein in the reporter cells. Images are false-colored. b, Stop codon readthrough in cells growing in liquid cultures. BL21 cells transformed with the pRXG[UGA] reporter plasmid were grown in liquid cultures (n=3 biologically independent samples ) in the presence of subinhibitory concentrations of Dro or Api with continuous monitoring of light absorbance (A600) and fluorescence of RFP and GFP proteins. The bar graph represents the ratios of GFP and RFP fluorescence values (relative fluorescence units, rfu) after 9 h of culture growth. For this experiment, the concentrations of PrAMPs at which no growth was detected after 9 h were taken as MIC (1 μM for Api and 4 μM for Dro). Error bars show standard deviation from the mean. Statistical significance of the difference from the no PrAMP control is shown (2-tailed t-test: n.s., not significant, p > 0.05; * 0.01< p ≤ 0.05; ** 0.001 < p ≤ 0.01; *** p ≤ 0.001). Individual p values are shown in the Source data file.
Endogenous expression of Dro is toxic for bacteria
Previous efforts to improve the antibiotic activity of Dro were based on chemical synthesis of peptides with specific amino acid changes 27,38,39. Although some alterations yielded derivatives with somewhat improved antibacterial properties or serum stability 38,40–42, labor- and time-consuming peptide synthesis limits the systematic evaluation of the contribution of the PrAMPs residues for activity. Furthermore, testing synthetic PrAMPs in whole-cell assays complicates distinguishing the effects of the structural alterations on cellular uptake versus on-target activity.
Because the non-glycosylated Dro remains highly activity (Figs. 1c and 2, Table 1), we decided to evaluate the structure-activity relationship (SAR) of Dro by expressing mutant variants directly in the bacterial cell. This approach, which has been successfully used for the analysis of several other antimicrobial peptides 7,43–49, alleviates the need for intracellular import and therefore reveals the contribution of individual amino acids to the action of the PrAMP upon the ribosome.
To test for the inhibitory activity of endogenously expressed Dro, its coding sequence was codon-optimized for expression in E. coli, equipped with start (AUG) and stop codons (UGA or UAG), and introduced into plasmids under the control of the L-arabinose (L-Ara) inducible PBAD promoter 7 (Fig. 3a). However, in contrast to the highly toxic expression of Api from similar constructs 7, Dro expression did not prevent the growth of E. coli BL21 cells (Extended Data Fig. 2). Because our in vitro assays showed that the ribosome-stalling activity of Dro was weaker than that of Api (Fig. 1c), we reasoned that toxicity of Dro could become evident in cells growing under less favorable conditions. Indeed, growth in a minimal (M9) medium rendered E. coli cells susceptible to the endogenous expression of Dro (Fig. 4).
Fig. 3: Endogenous expression of Dro is toxic for bacteria due to the interactions of the PrAMP with the ribosome and release factors.
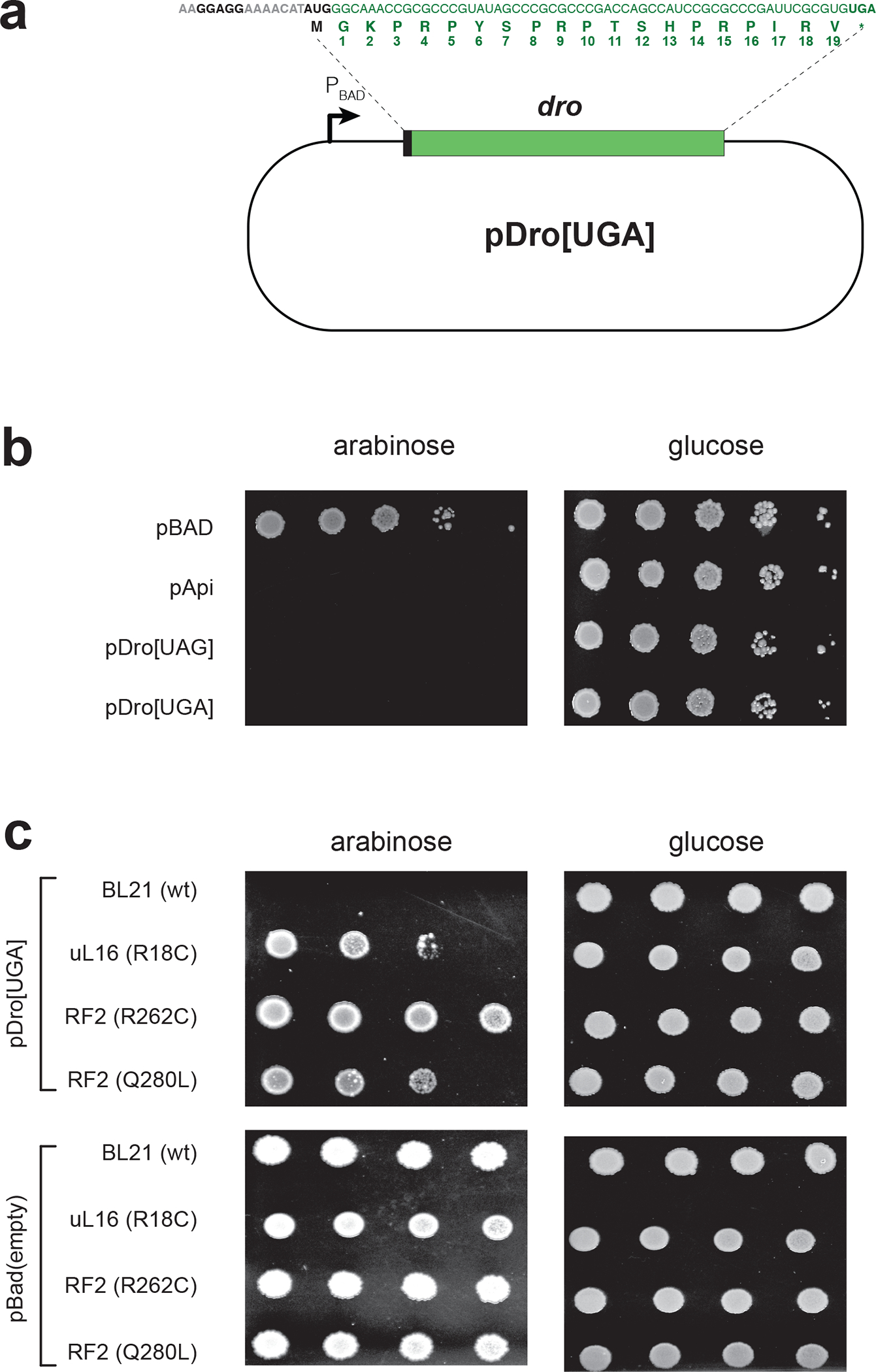
a, Schematics of the pDro[UGA] plasmid for expression of Dro in E. coli cells. The L-arabinose inducible PBAD promoter is indicated. The Dro encoding sequence, preceded by a ribosome binding site and a start codon, is shown in green. The start and stop codons are in bold. Numbering of the Dro residues, corresponding to the naturally produced PrAMP, is shown below the amino acid sequence. The sequence of the pDro[UAG] plasmid is identical except that the stop codon of the dro ORF is UAG. b,c, Spot test to determine the effect of endogenous Dro expression in (b) wt E. coli BL21 cells or (c) BL21 derivatives carrying mutations in ribosomal protein uL16 or RF2. Ten-fold (panel b) or three-fold (panel c) dilutions of cell cultures were spotted on M9 minimal medium agar plates under non-inducing (glucose) or inducing (L-Ara) conditions. Api-expressing cells (pApi) were used for comparison. Cells transformed with an empty pBad vector were used as control. The effect of Dro expression upon cells grown in rich medium is shown in Extended Data Fig. 2.
Fig. 4: The growth inhibition effect of Dro mutants expressed in cells.
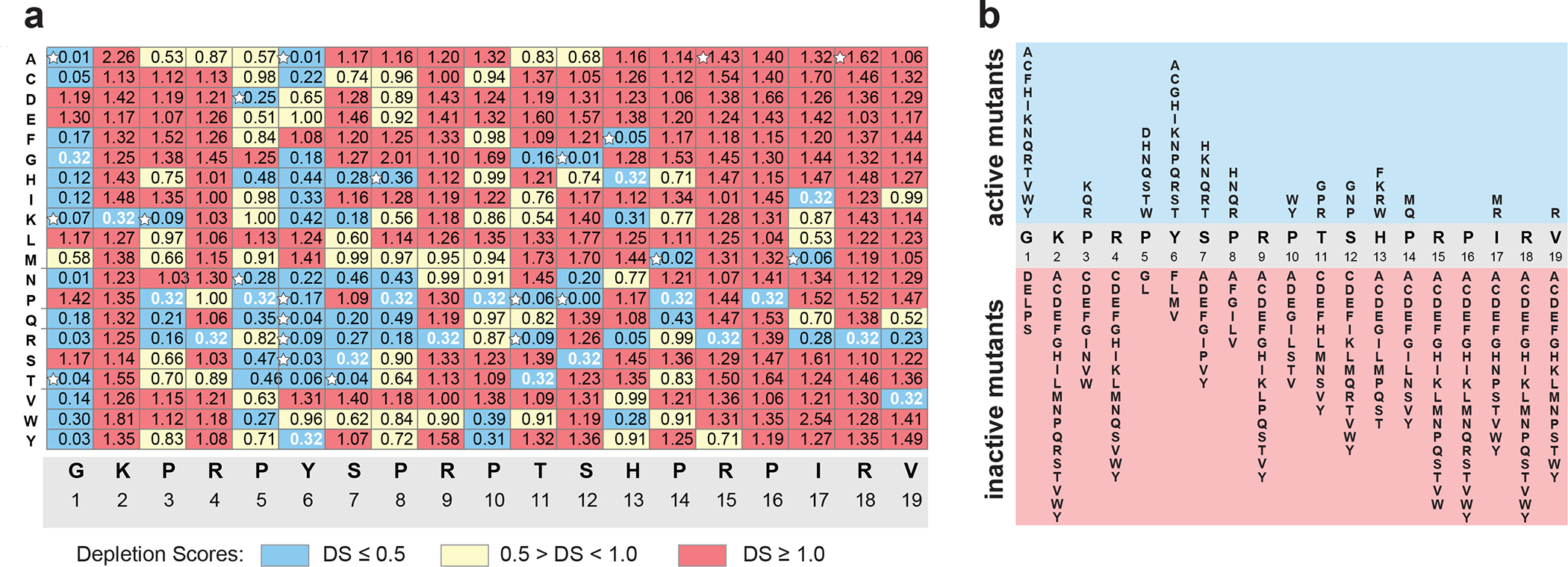
a, Depletion scores (DS) characterizing effects of single-amino acid substitutions upon the ability of endogenously expressed Dro variants to arrest cell growth. DS reflects the difference in abundance of a particular variant in the library of mutant dro genes in pDro[UAG] plasmids expressed in E. coli BL21 cells grown under inducing versus non-inducing conditions. Dro mutants were classified as highly toxic (DS ≤0.5) (blue), mildly toxic (DS 0.5 to 1.0) (yellow) or not toxic (DS ≥1) (red). DS value (0.32) of the wt Dro is shown with white numbers. Variants selected for chemical synthesis and further analysis are marked with white stars. The DS plot of the mutant library in pDro[UGA] plasmids is shown in Extended Data Fig. 3b. b, Diagram summarizing the effect of the Dro mutations on cell growth. Only highly toxic (blue field) and non-toxic (red field) mutants are shown.
It was important to verify that the toxicity of endogenously expressed Dro is due to its ability to interfere with translation termination. To this end, we once again exploited the Api- and, as we now know, Dro-resistant mutants with alterations in the ribosome (protein uL16) or in RF2 21 (Table 1). Dro-resistant mutants tolerated the endogenous expression of Dro much better than the parental wt strain (Fig. 3c). Therefore, we concluded that when translated in bacteria, Dro retains its ability to bind to the ribosome and interfere with the termination of translation.
Specific Dro residues are critical for on-target activity
Although Dro and Api show a convergent mode of action, they have significant differences in the amino acid sequences (Fig. 1b). Therefore, it was important to deduce which Dro residues are important for ribosome binding and translation inhibition. To achieve this goal, we assessed the contribution of each amino acid of the endogenously expressed Dro to inhibition of cell growth. We prepared two pDro plasmid libraries of mutant Dro genes encoding, besides the wt peptide, all possible single amino acid Dro mutants (n=19×19=361) and ending with either the UAG or UGA stop codons. The libraries were transformed into E. coli BL21 cells and plated at high density onto M9/agar plates supplemented with glucose (non-inducing conditions) or L-Ara (inducing conditions). After deep sequencing of the libraries, we computed the ‘depletion score’ (DS) for each mutant, which reflected the difference in its abundance in the libraries grown under inducing vs non-inducing conditions. Clones expressing inhibitory peptides are expected to be depleted upon induction and the DS values of such mutants should be less than 1. As a consequence, the relative abundance of clones expressing inactive Dro variants should increase on the L-Ara plate and the respective DS values should be above 1 (Fig. 4). Consistent with this metric, the DS value of wt Dro is 0.3. The results obtained with the libraries ending with UGA or UAG stop codons were highly convergent (Fig. 4a,b, Extended Data Fig. 3a,b), indicating the independence of the activity of the expressed Dro peptides from the nature of the stop codon of the dro gene and emphasizing the robustness of the obtained results.
The overall distribution of Dro-inactivating mutations shows a characteristic bias with many of them clustering towards the C-terminal segment of the PrAMP (Fig. 4; Extended Data Fig. 3b, chart cells colored red). In particular, the sequence encompassing Thr11-Val19 appears to be critical for the on-target activity, as most mutations of these residues inactivate the endogenously expressed Dro. Three additional residues in the N-terminal segment, Lys2, Arg4, and Arg9, are also pivotal for the activity: any mutation of these amino acids, including substitution with similarly positively charged residues, reduce Dro activity. In contrast, the Pro5-Pro8 segment and the N-terminal glycine can tolerate multiple mutations without any significant activity loss (Fig. 4; Extended Data Fig. 3b, chart cells colored blue). Thus, our mutational data strongly argue that an extended C-terminal segment of Dro as well as some of its residues proximal to the N-terminus are important for binding to the ribosome and trapping RFs. This pattern contrasts the results of mutational studies of Api, where only a few C-terminal amino acid residues were found to be functionally critical (Extended Data Fig. 3c), whereas the N-terminal sequence could be mutated or even deleted without significant impact on its activity 7. Thus, while sharing the general mechanism of action, different Type II PrAMPs utilize converging but clearly distinct strategies for target engagement.
Improved activity of specific Dro variants
Remarkably, the very low DS values of some of the endogenously expressed Dro mutants suggested that they may be more active than the wt non-glycosylated Dro (Fig. 4f Dro and tested their activity in vitro and in vivo (Fig. 4a, white stars). We also generated two high-DS-value peptides, R15A and R18A, to serve as negative controls (Fig. 4a).
Toeprinting analysis showed that the active mutant Dro variants are able to stall the ribosome at the UAG stop codon of the model ORF whereas the inactive control mutants failed to do so (Fig. 5a, Extended Data Fig. 4). In agreement with the activity charts (Fig. 4; Extended Data Fig. 3b), some of the synthetic peptides, including the S7T, T11P, T11R, S12G, H13F, P14M and I17M mutants, arrested translation more efficiently than the unmodified Dro (Fig. 5a). Strikingly, three of the tested mutants, T11P, T11R and particularly P14M, stalled the ribosome even better than Api137, which so far was known as the most active Type II PrAMP 7,21,24. These results suggest that the ribosome-targeting activity of non-glycosylated Dro can be significantly improved by small alterations in the peptide sequence.
Fig. 5: Synthetic Dro variants with single-amino acid substitutions preserve their ability to interfere with translation termination.
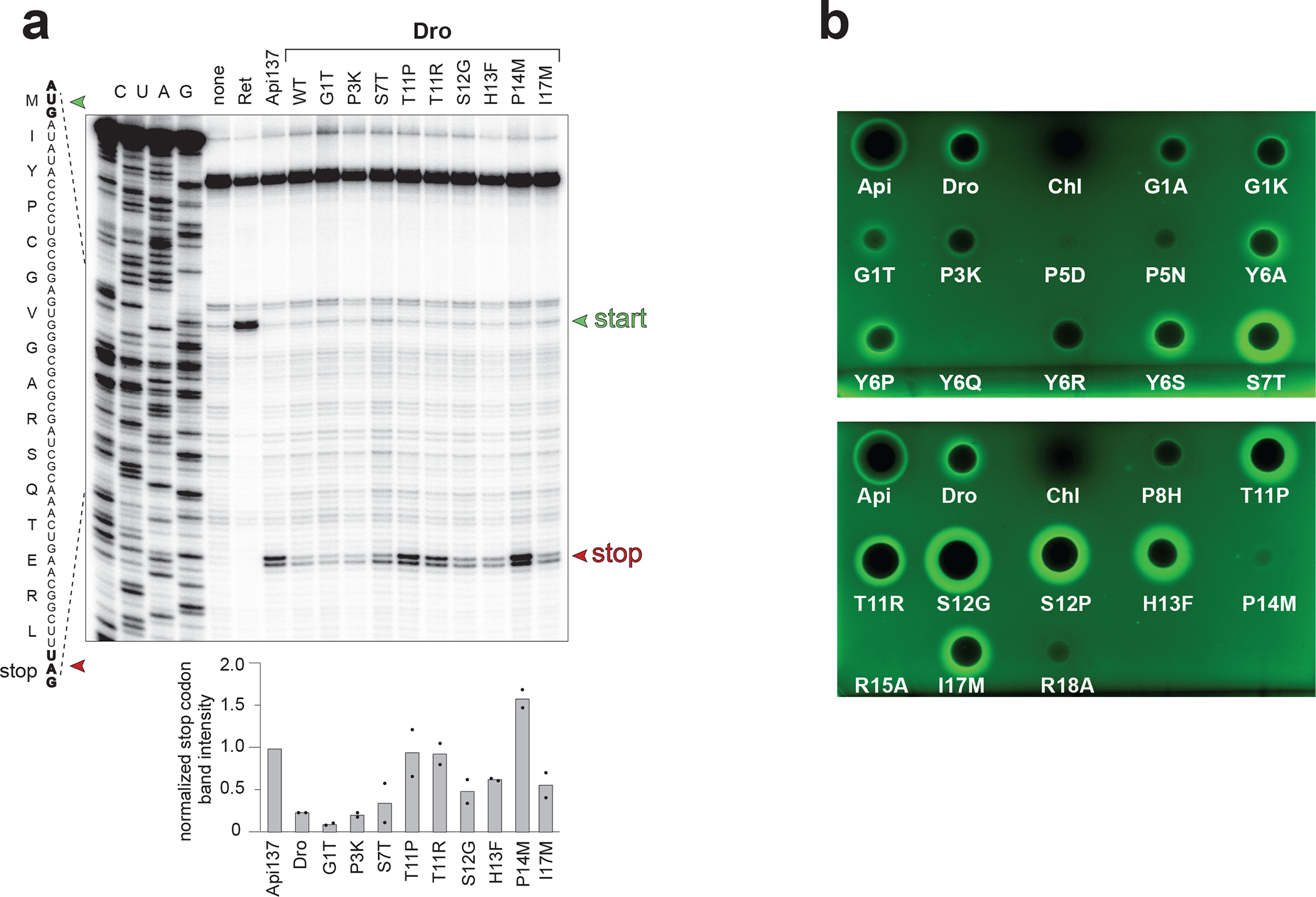
a, Toeprinting analysis of the ribosome arrest at the stop codon (red arrowhead) of the model ORF caused by synthetic wt or mutant non-glycosylated Dro variants. Arrest caused by Api137 was used as a positive control. The antibiotic retapamulin (Ret) was used to mark the toeprint band of ribosomes arrested at the start codon (green arrowhead). Bar graph shows the normalized average intensity of the toeprint bands of ribosomes arrested at the stop codon estimated from two independent experiments. Individual values from the two experiments are marked with dots. Intensity of the toeprint band generated by Api137-arrested ribosomes was set as 1.0. The complete toeprinting analysis of all the synthesized mutants is shown in Extended Data Fig. 4. b, Antibacterial (the clearing zones) and stop codon readthrough (the GFP fluorescence halos) activities of synthetic Dro variants in E. coli BL21 cells transformed with the pRXG[UGA] reporter plasmid. Api137 (1/10 amount relative to Dro variants) was used for comparison and Chl was used as a stop codon read-through negative control.
Efficient cellular uptake, facilitated by the SbmA-type peptide transporters, is required for the antibacterial activity of PrAMPs 10,11. Therefore, it was unclear whether any of the Dro mutants that stopped cell growth when expressed directly in bacteria would exhibit antibacterial properties when added exogenously to cells. Nevertheless, the MIC testing showed that most of the synthetic mutant PrAMPs exhibited the antibiotic properties comparable to those of the non-glycosylated wt Dro (Supplementary Table 1). Unexpectedly, the MIC of one of the negative control mutants, R18A, was only 4 times higher than that of the wt Dro (2 μg/mL), indicating that in contrast to the other negative control mutant, R15A, this PrAMP variant retained some antibacterial activity.
We further tested whether the synthetic Dro variants can induce stop codon readthrough. We used the drop diffusion assay with the E. coli BL21 cells carrying the RFP-GFP reporter (Fig. 2b) to simultaneously verify the cell-inhibitory activity of Dro mutants on solid medium and test their ability to induce stop codon bypass. In agreement with the MIC results, the majority of the tested mutants inhibited E. coli growth on agar plates and most of such mutants were also able to induce GFP expression due to leaky translation (Fig. 5b, Supplementary Table 1). The extent of GFP induction (Fig. 2b), generally matched the ability of the Dro variants to stall the ribosome at the stop codon in vitro (Fig. 5a). Consistently, the mutants S7T, T11R, T11P, S12G induced stop codon readthrough in vivo much more efficiently than wt Dro (Fig 5a, b).
Altogether, our data show that by introducing limited mutations into the structure of the unmodified Dro it is possible to significantly improve target engagement and possibly antimicrobial activity of this PrAMP without requiring synthetically challenging posttranslational glycosylation.
Discussion
In this work we examined the mechanism of action of the PrAMP Dro from fruit flies. We found that Dro interferes with bacterial growth by acting upon the ribosome and arresting it at stop codons likely trapping class 1 RFs in the post-release complex. Interrogating comprehensive libraries of single amino acid mutants, we identified Dro residues critical for activity. We further demonstrated that specific mutations could increase the ability of non-glycosylated Dro to arrest the terminating ribosome, stimulate stop codon readthrough, and possibly improve the antibacterial properties of the PrAMP.
The mechanism of Dro action resembles that of the only known Type II PrAMP, Api. However, Api and Dro employ notably different strategies for target engagement. Api requires only few of its C-terminal residues to bind tightly to the ribosome; mutations of most of the other amino acids do not interfere with the ability of endogenously expressed Api to inhibit cell growth 7 (Extended Data Fig. 3c). Consistently, the Api’s N-terminal segment is poorly resolved in cryo-EM reconstructions of the ribosome-Api137 complex, suggesting that it is disordered and forms only weak non-specific interactions with the NPET 21. In contrast, functionally important amino acid residues are distributed through the entire length of Dro and some of them (Lys2, Arg4, and Arg9) are found in the Dro’s N-terminal segment. This finding is consistent with the cryo-EM structure of the ribosome-Dro complex (see the accompanying paper of Koller et al. 50), where the entire peptide is well resolved due to the multiple interactions with the NPET. Our mutational data help to discern which of the contacts between Dro residues and the ribosome observed in the cryo-EM reconstructions of Koller at al. 50 are essential for the PrAMP’s activity and which may play only a secondary role (Extended Data Fig. 5). Any mutation of the Arg residues of Dro (Arg4, Arg9, Arg15, and Arg18) diminishes activity, indicating their importance for Dro action. Stacking of Arg9 side chain upon the nitrogen base of A751 and of Arg15 upon 23S rRNA A2062, as well as the hydrogen bonding of Arg9 with ribosomal protein uL22 and of Arg18 with RF, are likely pivotal for Dro action (Extended Data Fig. 5a–c). The reason for the Arg4 importance is less clear because its closest interacting partner, the non-bridging oxygen of the U1258 phosphate, is 4 Å away – too far to establish a strong hydrogen bond (Extended Data Fig. 5d). It is possible, however, that the lack of Thr11 glycosylation may cause a slight relocation of Arg4 in the NPET strengthening its interaction with U1258 making this contact particularly important for the non-glycosylated Dro. Two other ‘untouchable’ Dro residues are Lys2 and Pro16 (Fig. 4). The negative impact of the Lys2 mutations may point to the importance of the interaction of this residue with the G1256 phosphate (Extended Data Fig. 5e). In contrast, Pro16 does not form direct contacts with the ribosome; its invariance likely stems from the necessity to properly orient the neighboring Arg15 to contact A2062 and/or to position the peptide’s C-terminus for interactions with the RF (Extended Data Fig. 5a,b).
While in the cryo-EM model 50 Tyr6 and Ser7 form direct contacts with the ribosomal proteins uL4 and uL22 (Extended Data Fig. 5f), many mutations of these residues preserve Dro activity 40 (Fig. 4). Either their interactions contribute insignificantly to Dro binding or can be replaced with alternative interactions provided by the mutated residues.
The N-acetylglucosamine sugar at Thr11 of the native Dro forms specific interactions with the ribosome 50. It is possible, therefore, that some of the mutations that enhance potency of the unmodified Dro compensate for the lack of Thr11 modification. For example, Pro14 potentially imposes conformational constraints upon Dro’s trajectory facilitating the interaction of the sugar residue with 23S rRNA U2609. Such constraint, beneficial for binding of the glycosylated PrAMP, may become suboptimal when the sugar is absent which would explain why replacement of Pro14 with methionine may significantly stimulate the ribosome-arresting ability of non-glycosylated Dro (Fig. 5a; Extended Data Fig. 4).
While illuminating, the results of our mutational analysis with bacteria should be interpreted with a certain degree of caution. The activity of the endogenously expressed Dro could depend on its stability inside the cell, which can be affected by the mutations. It is also possible that alterations of the early codons of the dro gene may influence the efficiency of Dro expression rather than its activity. Nevertheless, it remains highly likely that toxicity of the endogenously expressed Dro variants directly reflects the ability of the PrAMPs to bind to the ribosome and inhibit translation.
The location and orientation of Type II PrAMPs in the RF-bound ribosome strikingly resemble the placement of the fully synthesized nascent protein after RF-promoted hydrolysis of peptidyl-tRNA. However, while completed proteins rapidly dissociate from the ribosome, Dro and Api establish long-lasting interactions with the NPET. Conceivably, endogenously expressed Type II PrAMPs may act in cis by remaining stuck in the NPET of the ribosome on which they were just synthesized. The possibility that wt Api, non-glycosylated Dro, and many of their multiple variants can remain within the NPET after their synthesis, suggests that the C-terminal segment of a wide spectrum of proteins may form strong enough interactions with the NPET to allow for the nascent protein to linger in the ribosome after separation from tRNA. Identifying such sequences in the cellular proteome could provide new insights into translation regulation, protein folding and targeting. The structural malleability of Type II PrAMPs also opens broad opportunities for optimizing their sequence and structure for medical applications.
The view that Type II PrAMPs can only inhibit translation termination and trap RFs on the ribosome could be too simplistic and may not capture the entire spectrum of the effects inflicted by Dro and its mutants in the bacterial cell. For example, while the synthetic mutant Dro peptides Tyr6Ser and Ser7Thr show comparable antibacterial activity, the Ser7Thr variant is more potent in stimulating GFP expression via stop codon readthrough (Fig. 5b). Similarly, bacterial growth is comparably inhibited by the Thr11Arg and Ser12Gly peptides, but the latter more efficiently causes stop codon readthrough (Fig. 5b). Possibly additional, more cryptic modes of action, e.g., arrest of initiation of translation of some genes, as was observed for Api 26, could contribute to the inhibitory activity of some of the Dro mutants.
In conclusion, our study has not only revealed the major mode of action of Dro and identified its residues critical for activity, but it also showed that by substituting only a few amino acids in the structure of a non-glycosylated PrAMP, it is possible to improve its on-target activity and possibly antibiotic properties. This may allow further optimization of Dro and related PrAMPs for potential therapeutic use bypassing the necessity for the experimentally challenging introduction of posttranslational modifications. Furthermore, expanding the variety of type II PrAMPs may yield new useful tools for studying fundamental mechanisms of translation termination and ribosome stalling.
Methods
Peptides and oligonucleotides.
Apidaecin 1b (Api1b) was synthesized by Genescript and Api137 was synthesized by NovoPro Biosciences, Inc., both at 95% purity. All the other peptides were synthesized in-house on solid phase as described 33. The DNA oligonucleotides libraries for generating Dro mutant libraries were ordered from Twist Biosciences. All other DNA oligonucleotides were from Integrated DNA Technologies.
In vitro translation and toeprinting analysis
The sf-GFP reporter protein was expressed essentially as described 33 in the PURExpress ΔRF123 system (New England Biolabs). The DNA template was PCR-amplified from the pY71sfGFP plasmid 51 by PCR using primers sfGFP-fwd and sfGFP-rev (all primers are listed in Supplementary Table 2). Transcription-translation reactions containing 35 ng of the DNA template were carried in a final volume of 5 μL. When needed, 0.5 μL of a 1:50 dilution of the RF1solution included in the kit were added to the reaction. Fluorescence of sfGFP was recorded in a microplate reader (Gemini EM, Molecular Devices) at 37°C for 2 h (λexc = 485 nm, λem = 535 nm). Fluorescence values used for Extended Data Fig. 1a correspond to the time point of 2 h.
Toeprinting experiments were carried out following the procedure described previously 21,52. The yrbA-fs15 DNA templates (with TAG or TGA stop codons), derived from the yrbA E.coli gene, used for in vitro transcription/translation reactions in the classic PURExpress system (New England Biolabs) were prepared by PCR, as previously described 7,21, by combining the overlapping primers T7, T7-IR-AUG, IR-yrbA-fs15-RF1 (for TAG) or IR- yrbA-fs15-RF2 (for TAG), post-NV1, and NV1, rendering the sequences TAATACGACTCACTATAGGGCTTAAGTATAAGGAGGAAAACATATGATATACCCCTGCGGAGTGGGCGCGCGATCGCAAACTGAACGGCTT(TAG/TGA)GCCGACCTCGACAGTTGGATTCACGTGCTGAATCCTGATGCGATGTCGAGTTAATAAGCAAAATTCATTATAACC (ORF sequence is underlined). PrAMPs or retapamulin were added from stock solutions in water to a final concentration of 50 μM.
MIC testing
Antimicrobial activity of Dro or its variants was determined by the MIC test in 96-well plates as described 7,33. Tests were performed with wild-type E. coli BL21(DE3) cells transformed with the pSbmA plasmid or its derivatives containing mutations in uL16 (Arg81Cys) or in RF2 (Arg262Cys or Gln280Leu) 7. MIC testing of synthetic Dro variants was carried out using untransformed BL21(DE3) cells. Testing the effects of dnaK knock-out was carried out in BW25113 strain or its dnaK::kan derivative from the Keio collection 53. Tests were performed in cell cultures grown at 37°C in 25% Cation-adjusted MHB medium. PrAMPs were serially diluted from stock solutions in water.
Stop codon readthrough assay
The ability of Dro or its variants to cause readthrough of premature stop codons in bacterial cells was tested by the dual RFP/GFP fluorescence pRXG plasmid reporter assay. E. coli BL21 cells were transformed with the original pRXG plasmid, where RFP and GFP encoding genes are separated by the TAG stop codon 37, or an engineered mutant version of the original pRXG plasmid where the stop codon was changed from TAG to TGA. Cell were grown overnight in LB medium supplemented with 50 μg/mL kanamycin (Kan) and then diluted 1:10 into fresh LB/Kan medium. Exponentially growing cells at an A600 of ~1 were pelleted and resuspended in the same volume of M9 minimal medium supplemented with 2 mM MgSO4, 0.1 mM CaCl2, 10 μg/mL thiamine. The cell suspension (3 mL) was poured over an agar plated prepared on supplemented M9 minimal medium and additionally containing 0.2 mM IPTG and 50 μg/mL Kan. After a brief (~30 s) incubation, excess liquid was aspirated, and plates were allowed to dry with lids removed. Solutions of Dro or its derivatives (2 mM), of Api137 (0.2 mM) (2 μL drops) or of 1 mg/mL of Chl (1 μL drop) were applied. Plates were incubated for 18 to 24 h at 37oC and imaged in a ChemiDoc Imaging System (BioRad) to detect RFP and GFP fluorescence (Cy3 and Cy2 channels, respectively). Images were processed and false-colored using the ImageLab software (Bio-Rad). The level of stop codon readthrough was quantified with the help of Image J by determining the total amount of GFP fluorescence above the background. For that first, the total fluorescence in the circle encompassing the visible zone of induction was determined. Then the fluorescence of the agar within the circle of no cell growth (clearing zone) was subtracted yielding the fluorescence of the ‘induction ring’. The fluorescence of the ring of the same size away from the PrAMP drop was taken as the background and was subtracted from the fluorescence value of the induction ring.
For the analysis of stop codon readthrough in liquid culture, E. coli BL21 cells transformed with pRXG[UGA] plasmid were gown at 37°C overnight in LB medium supplemented with 50 μg/mL Kan, then diluted into the same medium and grown to A600 ~0.4. Cultures were then diluted 1:100 into M9 medium supplemented with 0.4% glucose and 0.2 mM of IPTG, placed into wells of a 96-well (black-well clear-bottom) plate and varying concentrations of Dro or Api were added. The final culture volume was 100 μL per well. The PrAMP concentrations varied from 0 to 16 μM over two-fold dilutions. Cells were grown in the TECAN plate reader at 37 °C with monitoring every 20 min of cell density (A600), RFP fluorescence (λ ex = 550 nm, λ em = 675 nm, gain =100) and GFP fluorescence (λ ex = 485 nm, λ em = 520 nm, gain =125). Data were plotted and data points corresponding to 9 h of growth were used for calculating the GFP/RFP (RFU/RFU) ratios.
Endogenous expression of Dro in bacterial cells
The sequence of the DNA templates (synthesized by IDT) coding for the mature Dro peptide (GKPRPYSPRPTSHPRPIRV) was codon-optimized for expression in E. coli and ATG start codon and stop codons TAG or TGA (underlined in the sequence provided below) were added: ATGGGCAAACCGCGCCCGTATAGCCCGCGCCCGACCAGCCATCCGCGCCCGATTCGCGTGTAG (or TGA)
The Dro-encoding templates were used to swap the Api encoding gene with the Dro sequence in the pApi vector 7. For this, the Dro templates were introduced by Gibson assembly into pApi cut with SacI and XbaI restriction enzymes, yielding pDro[UAG] and pDro[UGA] plasmids.
To test the effects of the endogenously expressed Dro on cell growth, the pDro[UAG] and pDro[UGA] plasmids were transformed into E. coli BL21 cells. Transformants were grown overnight in LB medium supplemented with 30 μg/ml Chl and 2% glucose. Cultures were diluted 100-fold into fresh medium, grown at 37°C, and upon reaching an A600 ~0.5 they were spun and resuspended in LB or M9 medium. Ten-fold serial dilutions were spotted (3 μl) on LB/agar or supplemented M9/agar plates containing 30 μg/ml Chl, and either 2% glucose (non-inducing conditions) or 2% L-Ara (inducing conditions). The plates were incubated at 37°C for 16–48 h and photographed.
Spot-test
E. coli cells (BL21 or the mutant strains) transformed with pDro or pApi plasmids or with the empty pBad vector were grown overnight in LB medium supplemented with 30 μg/mL Chl and 0.4% glucose. Cultures were diluted 1:100 into M9 medium supplemented with 2% glycerol, 2 g/L tryptone and 30 μg/mL Chl and grown to 107 – 108 cfu/mL. Three μL of ten-fold (Fig. 2b) or three-fold (Fig. 2c) dilutions were spotted on M9 plates supplemented with 2% glycerol, 2 g/L tryptone, 30 μg/mL Chl, and containing either 0.4% glucose or 0.2 % L-arabinose. Plates were grown 36–48 h and photographed.
Mutant Dro library construction and selection screening.
Generating plasmid libraries.
Two libraries of the Dro genes, one ending with the TGA stop codon and another ending with the TAG codon were generated by massively parallel oligonucleotide synthesis. Each of the codons of the Dro-encoding gene was individually replaced by one of the 19 codons (with the highest E. coli codon adaptation index value) specifying each of the alternative amino acid residues. The dsDNA fragments (wt + 361 mutant for each library) of 363 nts each were procured from Twist Bioscience. The libraries were cloned into SacI/XbaI cut pBAD vector via Gibson Assembly, transformed into XL1-blue cells and plated on LB/agar plates supplemented with 30 μg/ml of Chl and 2% glucose yielding ~4×105 clones per library (>100-fold coverage). The clones were washed off the plates and the total plasmid was extracted using High-pure plasmid isolation kit (Roche).
Library selection.
The amplified plasmid libraries were then transformed into E. coli BL21 cells by electroporation and plated on ∅ 136 mm M9/agar plates containing 30 μg/mL Chl, 1% glycerol, and either 2% glucose or 2% L-Ara. The concentration of L-Ara used in the selection experiment corresponded to that required for strong inhibition of the growth of cells transformed with the original pDro[UAG] plasmid. Plates were incubated at 37° for 48 h. The clones (~4×105 per condition) were washed off the plates with 15% (v/v) glycerol, flash-frozen in liquid nitrogen and stored at −80°C.
Generating sequencing libraries.
Frozen glycerol stocks of libraries washed off the glucose- or L-Ara plates were thawed, and total plasmid was isolated using High-pure plasmid isolation kit (Roche). The dro genes were minimally PCR amplified (10 cycles) using Dro-Lib-Fwd and Dro-Lib-Rev primers. The PCR products were isolated using a DNA Clean and Concentrator kit (Zymo Research). Another round of PCR amplification (10 cycles) with the primers IDT8_i57 – IDT8_512 and IDT8_i77 – IDT8_i712 was used to add Illumina sequencing adapters to the libraries from each condition. The PCR products were size selected in a 15% TBE-Urea gel followed by elution, ethanol precipitation, and quantification in a Qubit fluorometer. The libraries were spiked with 30% PhiX due to low complexity and sequenced on an Illumina MiSeq platform at the DNA sequencing facility of Northwestern University.
Sequence analysis and phenotypic scoring.
Cutadapt (V3.4) was used to remove all the extra nucleotides from the 3’ and 5’ ends of the reads, leaving only the Dro sequence variants including the start and stop codons. Next, all the reads were sorted and counted based on unique sequence identity using the Unix command:
$ gzip -dc input_file.fastq.gz | paste - - - - | cut -f2 | sort | uniq -c | sort -g > output_file.txt Only exact matches to expected library sequences were considered for further analysis leaving a total of 432,013/179,563 sequences (glucose and arabinose conditions, respectively) for the UAG library and 163,449/295,374 sequences (glucose and L-Ara conditions, respectively) for the UGA library. The relative abundance of each mutant was calculated by the dividing the number of occurrences of its sequence by the total number of sequences in the library. Depletion score (DS) was then computed as log2 of the ratio of the relative abundance of the variant sequence in the induced (L-Ara) sample to the relative abundance of the same variant in the uninduced (glucose) samples. Further binning was as follows: DS values ≤ 0.5 - active Dro mutants; DS values > 0.5 but < 1.0 – moderately active mutants; DS values ≥ 1.0 – inactive mutants. The DS charts (Fig. 3; Extended Data Fig. 3) were generated in Excel.
Extended Data
Extended Data Fig. 1. Dro acts upon translation termination but causes weak translation arrest at UGA stop codons.
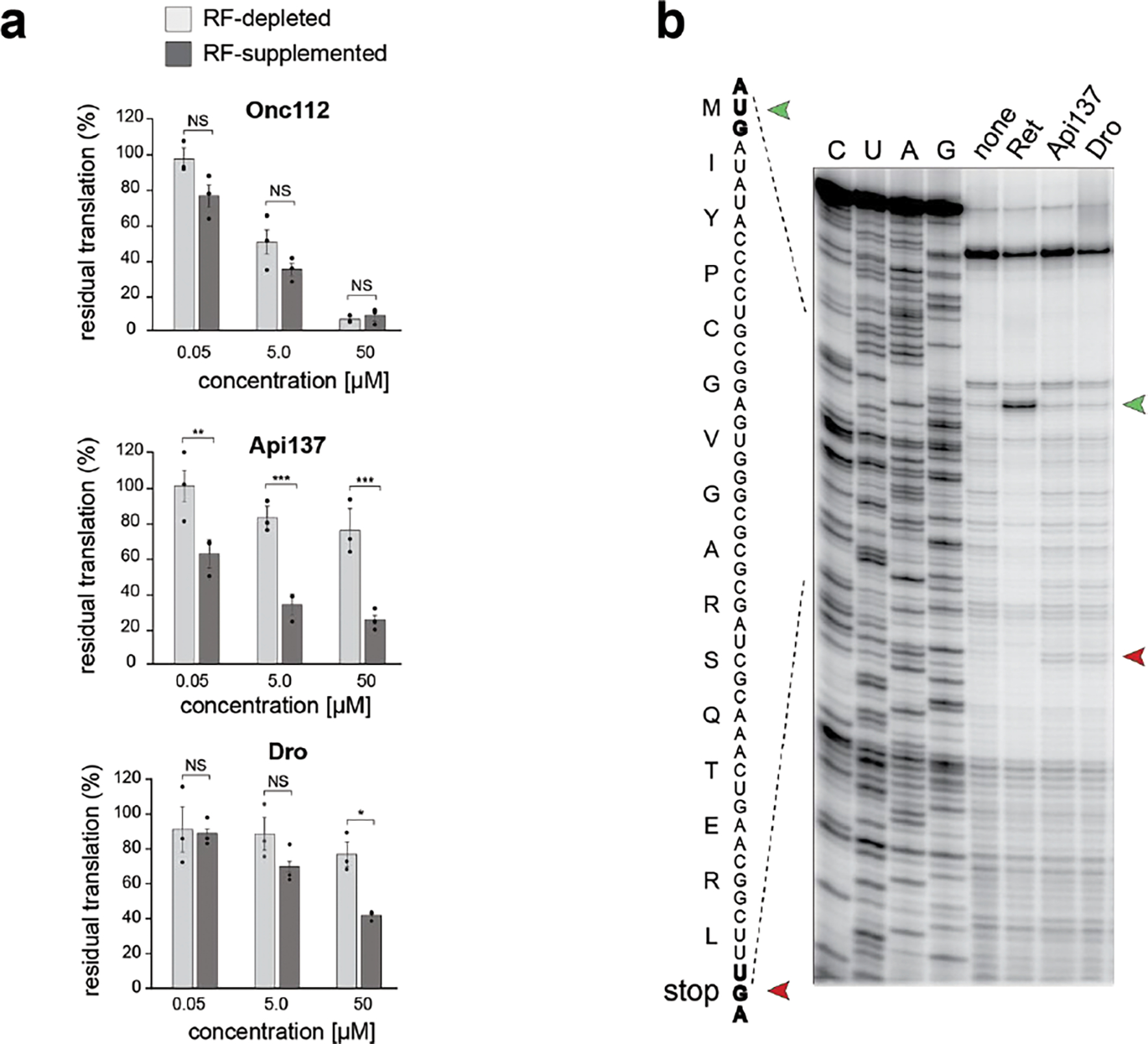
a, Inhibition of in vitro GFP translation by synthetic PrAMPs: class-I Oncocin112 (Onc112), class-II Api137, or non-glycosylated Dro (Dro). Bar graphs represent the normalized values of GFP fluorescence from reactions where RF1 was depleted (light grey) or supplemented (dark grey), setting the fluorescence value from reactions with or without RF1 in the absence of PrAMP as 100%. The error bars show standard deviation from the mean in three independent experiments. Significance levels indicated as NS, not significant; *, p-value <0.05; **, p-value < 0.01; ***, value < 0.001 (One-way ANOVA with Tukey’s Multiple Comparison test by GraphPad Prism). b, In vitro toeprinting analysis of the Api137 or Dro-mediated ribosome arrest at the UGA stop codon (red arrowhead) of the model yrbA ORF. The control reaction with no added PrAMPs is labeled as “none”. The control antibiotic retapamulin (Ret) stalls ribosomes at the start codon (green arrowhead). Sequencing reactions are labeled as C, U, A, G.
Extended Data Fig. 2. Endogenous expression of Dro in cells grown in rich medium does not prevent cell growth.
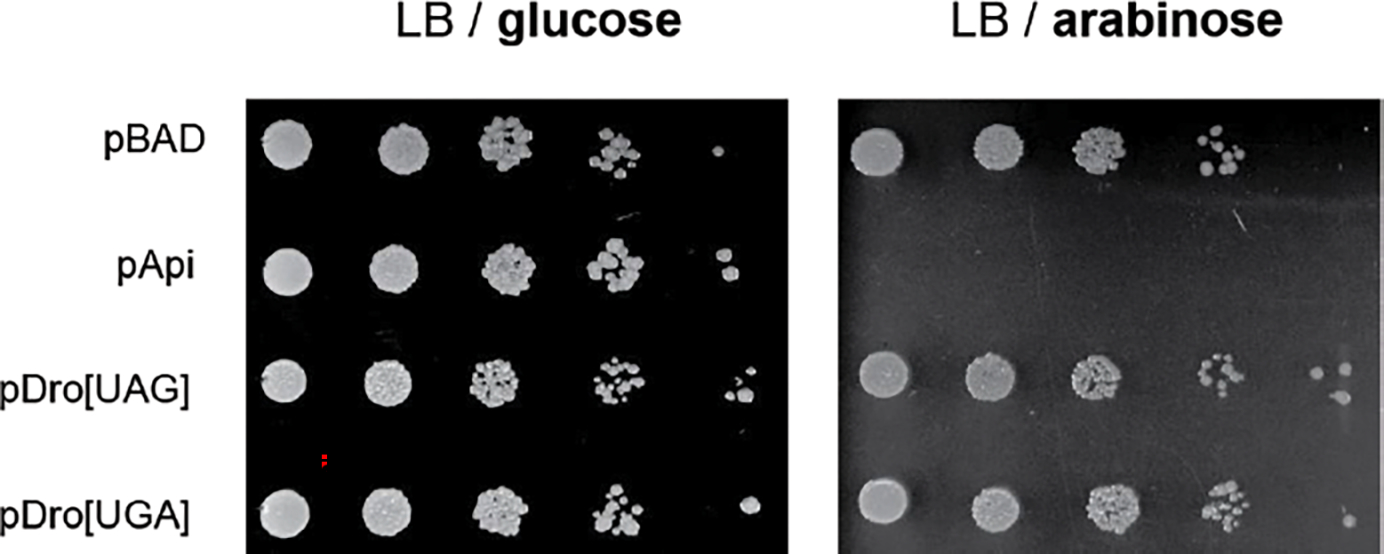
Growth of E. coli BL21 cells transformed with pDro[UAG] or pDro[UGA] plasmids on agar lysogeny broth (LB) rich medium supplemented with glucose or L-arabinose. The toxic effect of endogenous expression of Api (in cells transformed with pApi) is shown for comparison. Cells transformed with empty pBAD vector were used as a negative control.
Extended Data Fig. 3. Robustness of experimental data and comparisons of the effect of endogenously expressed Dro and Api variants on cell growth.
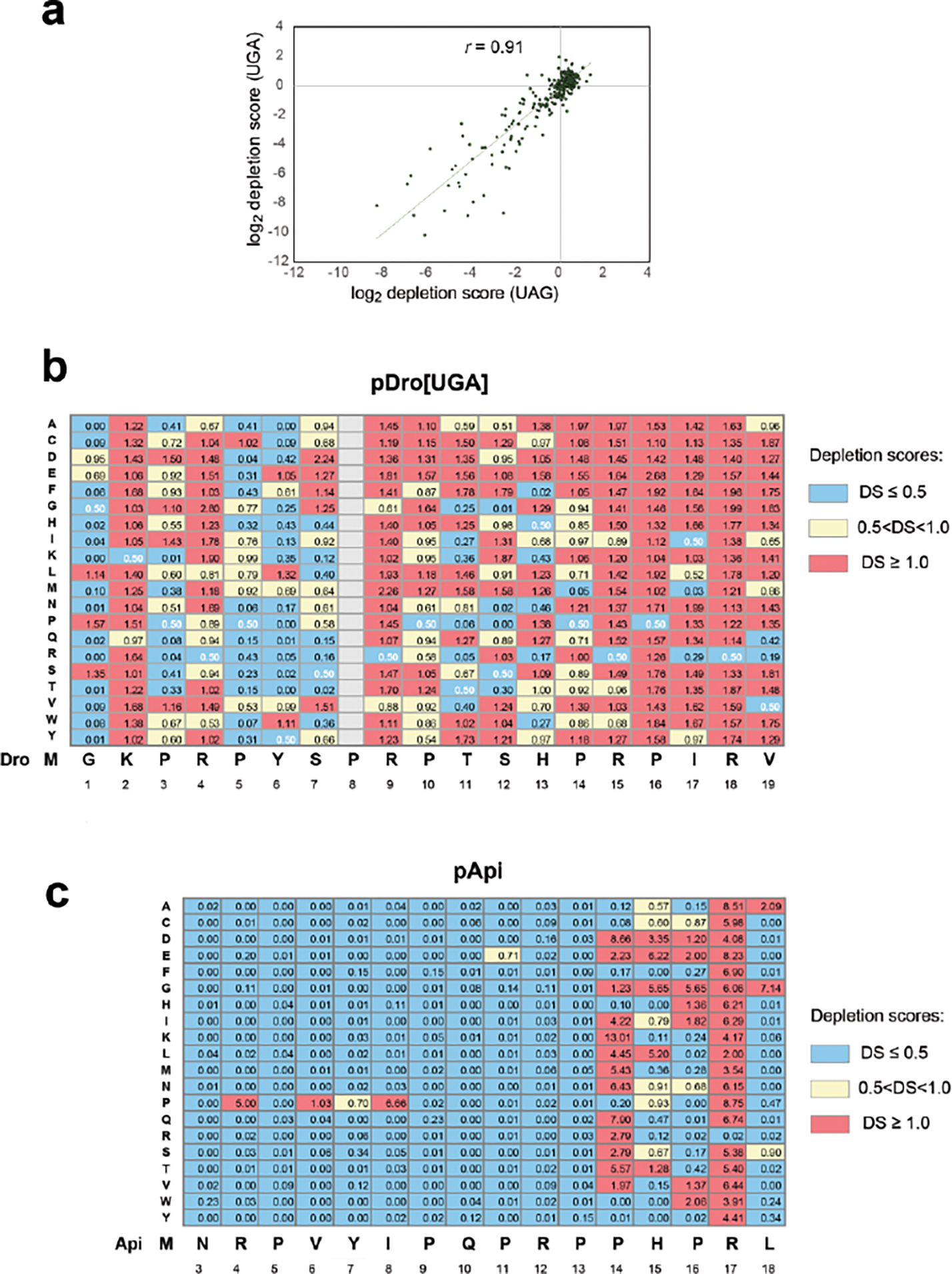
a, Correlation of the depletion scores of E. coli clones from the single amino acid Dro mutant libraries generated on the bases of pDro[UAG] or pDro[UGA] expression plasmids. Pearson correlation coefficient (r) is indicated. b, Depletion scores of library clones endogenously expressing single-amino acid Dro variants from the pDro[UGA] plasmids. Coloring is according to the toxic effects (blue - highly toxic, yellow - mildly toxic, salmon - not toxic). The analogous data for the pDro[UAG] library are shown in Fig. 3. c, Similarly calculated depletion scores for endogenously expressed Api mutants from the pApi plasmid using data from the reference 7.
Extended Data Fig. 4. Dro variants with single amino acid substitutions retain the ability to arrest ribosomes at stop codons.
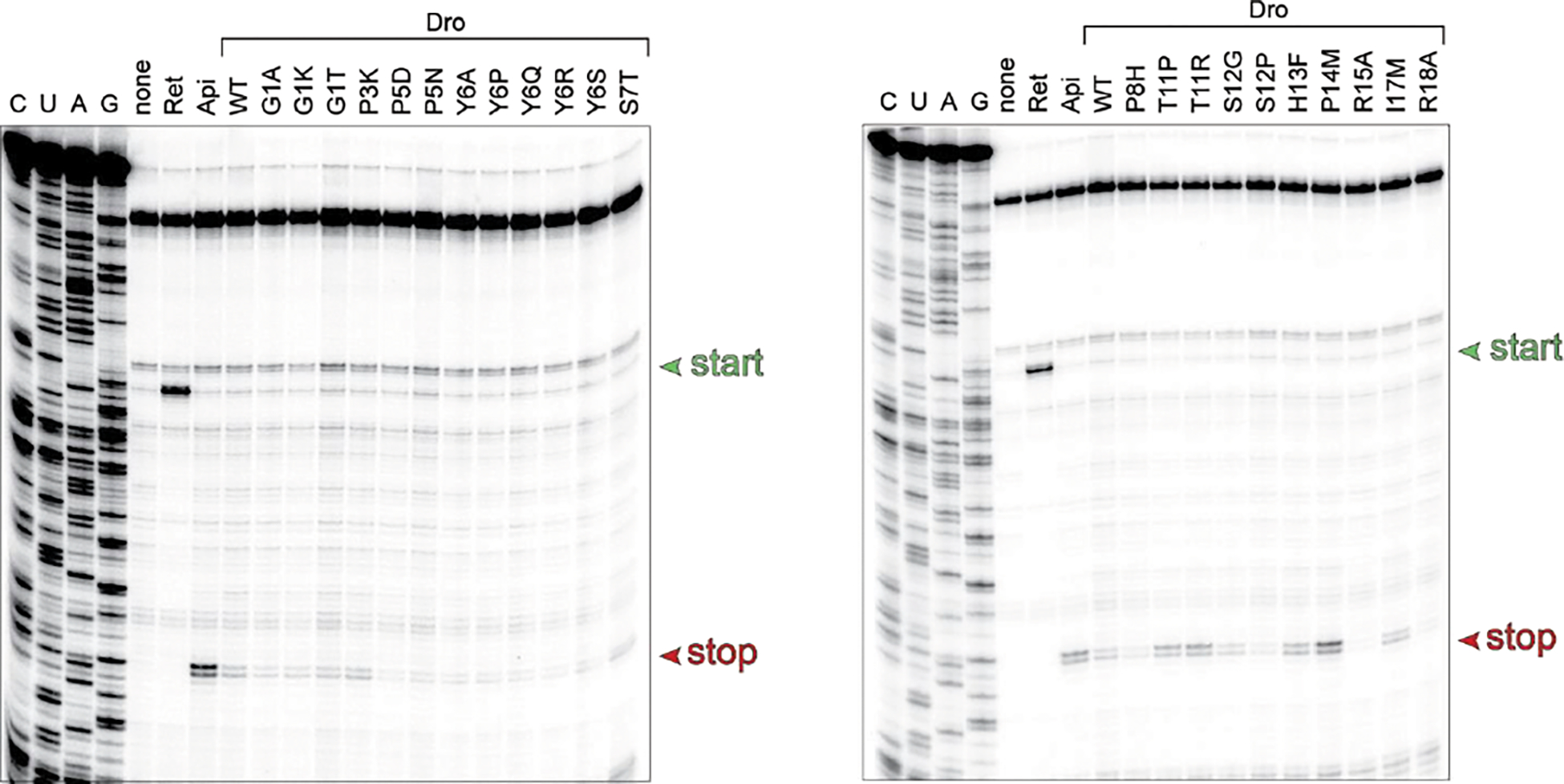
Toeprinting analysis of the ribosome arrest of the UAG stop codon of the model yrbA ORF mediated by synthetic non-glycosylated Dro variants. Samples with no added synthetic peptide are labeled as ‘none’. Arrest at stop codons caused by Api137 or at start codons by retapamulin (Ret) are shown as reference. Toeprint bands from ribosomes stalled at start or stop codons are marked by green and red arrowheads, respectively. Shown are representative gels of two independent experiments that produced converging results.
Extended Data Fig. 5. Functionally critical contacts of Dro with the ribosome as revealed by mutational analysis.
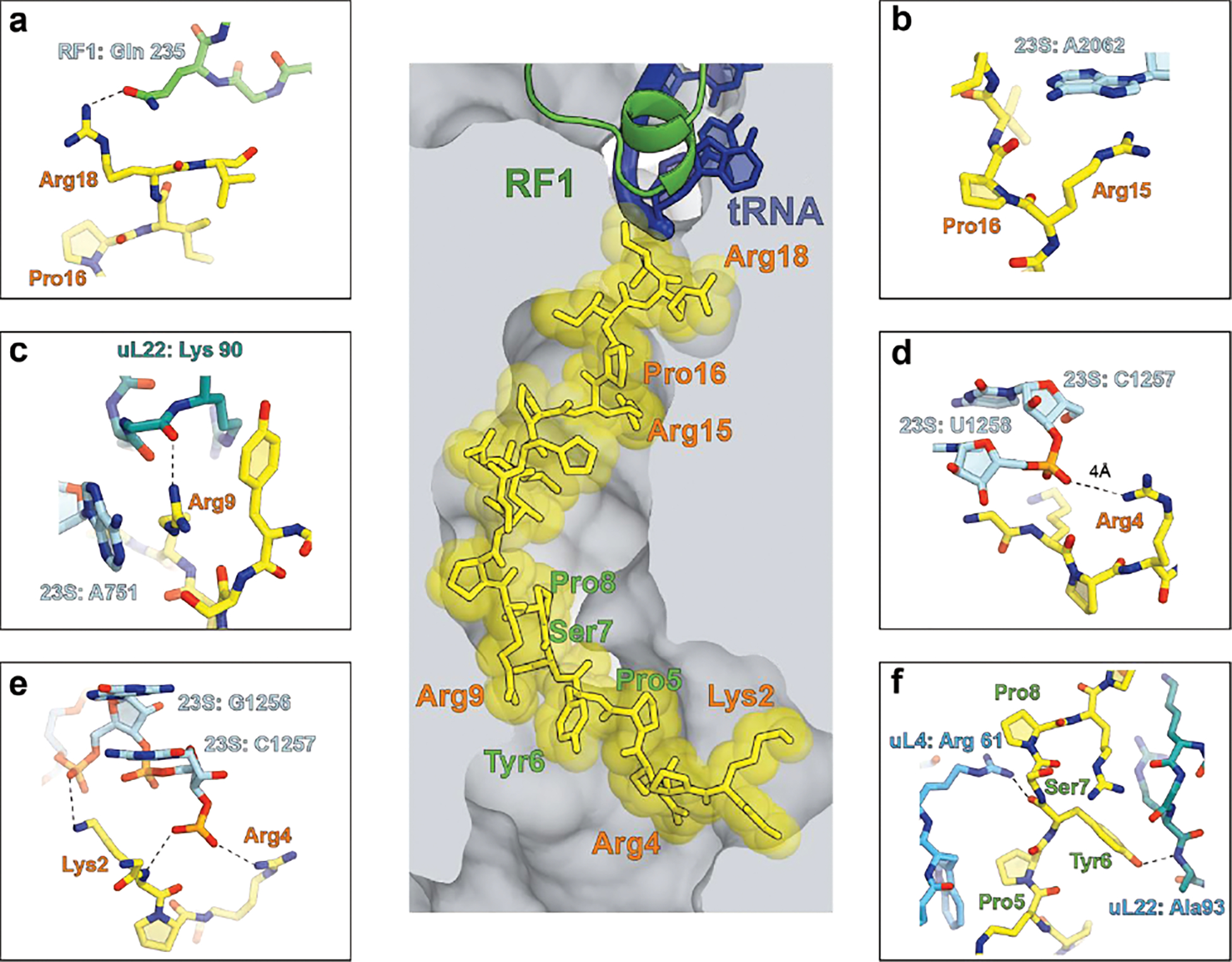
The central image depicts the placement of glycosylated Dro in the NPET of the E. coli ribosome 50. Functionally critical Dro residues are indicated in salmon; residues that tolerate multiple substitutions are marked in green. a-e, Functionally critical contacts involving Dro residues: a, Arg18, b, Arg15 and Pro16, c, Arg9, d, Arg4, e, Lys2. Panel f shows the ribosomal contacts of the Pro5-Pro8 segment of Dro where most amino acids substitutions do not impair the PrAMP’s activity.
Supplementary Material
Acknowledgements
We thank Timm Koller and Daniel Wilson (University of Hamburg) for generously sharing their results and the structure of the ribosome-drosocin complex. This work was supported in part by NIH grant R01 AI162961 (to ASM, NV-L and YSP).
Footnotes
Competing interests statement
Up until one year prior to submission of this manuscript, R.H. served as an advisor for the company EnBiotix, Inc. on a project unrelated to this study. The remaining authors declare no competing interests.
Data availability
The uncropped gels, the raw data used for calculating the Depletion Score and statistics data for Fig. 2b and ED Fig. 1a are shown in the Source Data file associated with the manuscript. Because raw sequencing data for the mutant Dro gene libraries do not represent genomic, RNA-seq or Ribo-seq results, they were not deposited to the public databases but are available from the corresponding authors upon request.
References
- 1.Lazzaro BP, Zasloff M & Rolff J Antimicrobial peptides: Application informed by evolution. Science 368, eaau5480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardoso MH et al. Non-lytic antibacterial peptides that translocate through bacterial membranes to act on intracellular targets. Int. J. Mol. Sci. 20, 4877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otvos L Jr. The short proline-rich antibacterial peptide family. Cell. Mol. Life Sci. 59, 1138–1150 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graf M & Wilson DN Intracellular antimicrobial peptides targeting the protein synthesis machinery. Adv. Exp. Med. Biol. 1117, 73–89 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Scocchi M, Tossi A & Gennaro R Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life. Sci. 68, 2317–2330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benincasa M et al. The proline-rich peptide Bac7(1–35) reduces mortality from Salmonella typhimurium in a mouse model of infection. BMC Microbiol. 10, 178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baliga C et al. Charting the sequence-activity landscape of peptide inhibitors of translation termination. Proc. Natl. Acad. Sci. USA 118, e2026465118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulet P et al. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 268, 14893–14897 (1993). [PubMed] [Google Scholar]
- 9.Cociancich S et al. Novel inducible antibacterial peptides from a hemipteran insect, the sap-sucking bug Pyrrhocoris apterus. Biochem. J. 300 (Pt 2), 567–575 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattiuzzo M et al. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Molec. Microbiol. 66, 151–163 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Krizsan A, Knappe D & Hoffmann R Influence of the yjiL-mdtM gene cluster on the antibacterial activity of proline-rich antimicrobial peptides overcoming Escherichia coli resistance induced by the missing SbmA transporter system. Antimicrob. Agents Chemother. 59, 5992–5998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otvos L Jr. et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39, 14150–14159 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Scocchi M, Lüthy C, Decarli P, Mignogna G, Christen P, Gennaro R The proline-rich antibacterial peptide Bac7 binds to and inhibits in vitro the molecular chaperone DnaK. Int. J. Pept. Res. Ther. 15, 147–155 (2009). [Google Scholar]
- 14.Czihal P et al. Api88 is a novel antibacterial designer peptide to treat systemic infections with multidrug-resistant Gram-negative pathogens. ACS Chem. Biol. 7, 1281–1291 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Krizsan A et al. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew. Chem. Int. Ed. Engl. 53, 12236–12239 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Krizsan A, Prahl C, Goldbach T, Knappe D & Hoffmann R Short proline-rich antimicrobial peptides inhibit either the bacterial 70S ribosome or the assembly of its large 50S subunit. Chembiochem 16, 2304–2308 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Seefeldt AC et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Molec. Biol. 22, 470–475 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Roy RN, Lomakin IB, Gagnon MG & Steitz TA The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Molec. Biol. 22, 466–469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagnon MG et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 44, 2439–2450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seefeldt AC et al. Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 44, 2429–2438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florin T et al. An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome. Nat. Struct. Molec. Biol. 24, 752–757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver J, Mohammad F, Buskirk AR & Storz G Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, e02819–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng S et al. Mechanism of actions of Oncocin, a proline-rich antimicrobial peptide, in early elongation revealed by single-molecule FRET. Protein Cell 9, 890–895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthold N et al. Novel apidaecin 1b analogs with superior serum stabilities for treatment of infections by Gram-negative pathogens. Antimicrob. Agents Chemother. 57, 402–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf M et al. Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1. Nat. Commun. 9, 3053 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangano K et al. Genome-wide effects of the antimicrobial peptide apidaecin on translation termination in bacteria. eLife 9, e62655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulet P, Urge L, Ohresser S, Hetru C & Otvos L Jr. Enlarged scale chemical synthesis and range of activity of drosocin, an O-glycosylated antibacterial peptide of Drosophila. Eur. J. Biochem. 238, 64–69 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Bikker FJ et al. Evaluation of the antibacterial spectrum of drosocin analogues. Chem. Biol. Drug. Des. 68, 148–153 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Gobbo M et al. Antimicrobial peptides: synthesis and antibacterial activity of linear and cyclic drosocin and apidaecin 1b analogues. J. Med. Chem. 45, 4494–4504 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Vonkavaara M et al. Francisella is sensitive to insect antimicrobial peptides. J. Innate. Immun. 5, 50–59 (2013). 10.1159/000342468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann R, Bulet P, Urge L & Otvos L Jr. Range of activity and metabolic stability of synthetic antibacterial glycopeptides from insects. Biochim. Biophys. Acta 1426, 459–467 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Hanson MA, Kondo S & Lemaitre B Drosophila immunity: the Drosocin gene encodes two host defence peptides with pathogen-specific roles. Proc. Biol. Sci. 289, 20220773 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig T, Krizsan A, Mohammed GK & Hoffmann R Antimicrobial activity and 70S ribosome binding of apidaecin-derived Api805 with increased bacterial uptake rate. Antibiotics 11, 430 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orelle C et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 57, 5994–6004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uno M, Ito K & Nakamura Y Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie 78, 935–943 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Kragol G et al. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016–3026 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Monk JW et al. Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli. ACS Synth. Biol. 6, 45–54 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Knappe D, Cassone M, Nollmann FI, Otvos L & Hoffmann R Hydroxyproline substitutions stabilize non-glycosylated drosocin against serum proteases without challenging its antibacterial activity. Protein Pept. Lett. 21, 321–329 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Lele DS, Dwivedi R, Kumari S & Kaur KJ Effect of distal sugar and interglycosidic linkage of disaccharides on the activity of proline rich antimicrobial glycopeptides. J Pept Sci 21, 833–844 (2015). [DOI] [PubMed] [Google Scholar]
- 40.de Visser PC et al. Biological evaluation of Tyr6 and Ser7 modified drosocin analogues. Bioorg. Med. Chem. Lett. 15, 2902–2905 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Lele DS, Talat S, Kumari S, Srivastava N & Kaur KJ Understanding the importance of glycosylated threonine and stereospecific action of Drosocin, a Proline rich antimicrobial peptide. Eur. J. Med. Chem. 92, 637–647 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Ahn M et al. Substitution of the GalNAc-alpha-O-Thr(1)(1) residue in drosocin with O-linked glyco-peptoid residue: effect on antibacterial activity and conformational change. Bioorg. Med. Chem. Lett. 21, 6148–6153 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Taguchi S, Mita K, Ichinohe K & Hashimoto S Targeted engineering of the antibacterial peptide apidaecin, based on an in vivo monitoring assay system. Appl. Environ. Microbiol. 75, 1460–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taguchi S, Nakagawa K, Maeno M & Momose H In vivo monitoring system for structure-function relationship analysis of the antibacterial peptide apidaecin. Appl. Environ. Microbiol. 60, 3566–3572 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taguchi S, Ozaki A, Nakagawa K & Momose H Functional mapping of amino acid residues responsible for the antibacterial action of apidaecin. Appl. Environ. Microbiol. 62, 4652–4655 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taboureau O et al. Design of novispirin antimicrobial peptides by quantitative structure-activity relationship. Chem. Biol. Drug Des. 68, 48–57 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Muthunayake NS et al. Expression and in vivo characterization of the antimicrobial peptide oncocin and variants binding to ribosomes. Biochemistry (2020). [DOI] [PubMed] [Google Scholar]
- 48.Kuru E et al. Release factor inhibiting antimicrobial peptides improve nonstandard amino acid incorporation in wild-type bacterial cells. ACS Chem. Biol. 15, 1852–1861 (2020). [DOI] [PubMed] [Google Scholar]
- 49.DeJong MP et al. A platform for deep sequence-activity mapping and engineering antimicrobial peptides. ACS Synth. Biol. 10, 2689–2704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koller TO, Morici M, Berger M, Safdari H, Lele DS, Beckert B, Kaur KJ, Wilson DN Structural basis for translation inhibition by the glycosylated antimicrobial peptide Drosocin from Drosophila melanogaster. XXX, XX–XX (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods section references
- 51.Bundy BC & Swartz JR Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug. Chem. 21, 255–263 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Orelle C et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic acids research 41, e144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The uncropped gels, the raw data used for calculating the Depletion Score and statistics data for Fig. 2b and ED Fig. 1a are shown in the Source Data file associated with the manuscript. Because raw sequencing data for the mutant Dro gene libraries do not represent genomic, RNA-seq or Ribo-seq results, they were not deposited to the public databases but are available from the corresponding authors upon request.


