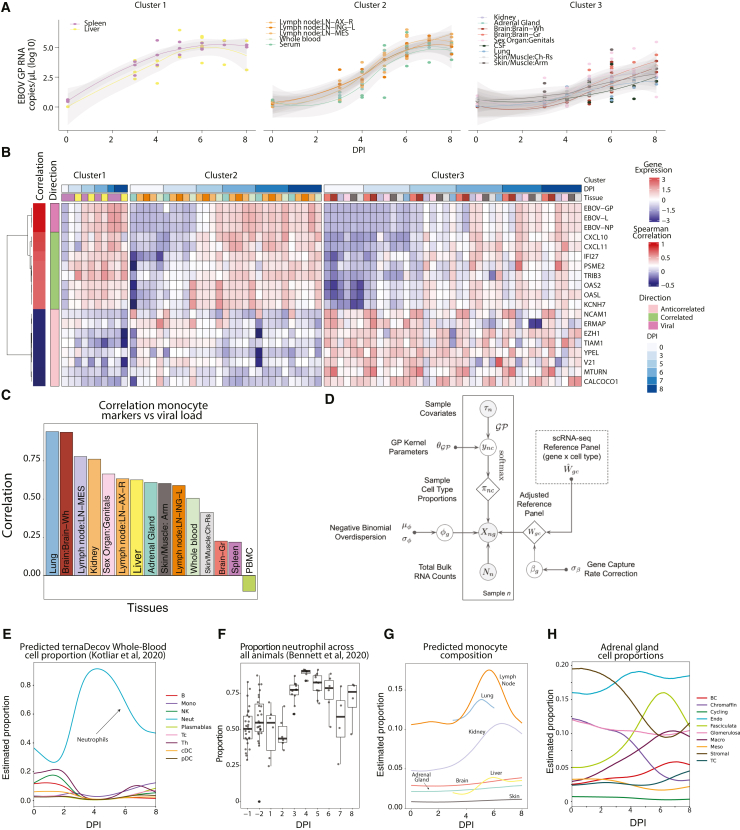
Figure 2.
Correlating viral dynamics and host response to infection
(A) Viral loads, as determined by qRT-PCR, plotted versus time. The trajectories for different tissues were separated into three distinct patterns using K-means longitudinal data clustering, yielding groups of tissues with similar viral load dynamics.
(B) Gene expression across tissues (separated by the clusters in A) for the top 8 correlated and anti-correlated DEGs and 3 representative viral genes. Samples are ordered along the x axis by tissue and DPI. On the y axis, DEGs are clustered and labeled by direction.
(C) Correlation between viral load and canonical monocyte marker expression across each tissue.
(D) Overview of modular deconvolution framework used in ternaDecov. The output proportions from the models are then used to draw observed sample counts from a negative binomial distribution based on the provided single-cell profiles.
(E) Deconvolution of whole blood using scRNA-seq data18 confirms the detected increase in neutrophils at 4 DPI.
(F) Proportion of neutrophils across samples using Sysmex XT-2000iV automated hematology by flow cytometry28.
(G) Deconvolution of monocyte composition across time for each tissue based on an scRNA-seq reference of Macaca fascicularis.
(H) Deconvolution of predicted cell type proportion across time for adrenal glands.