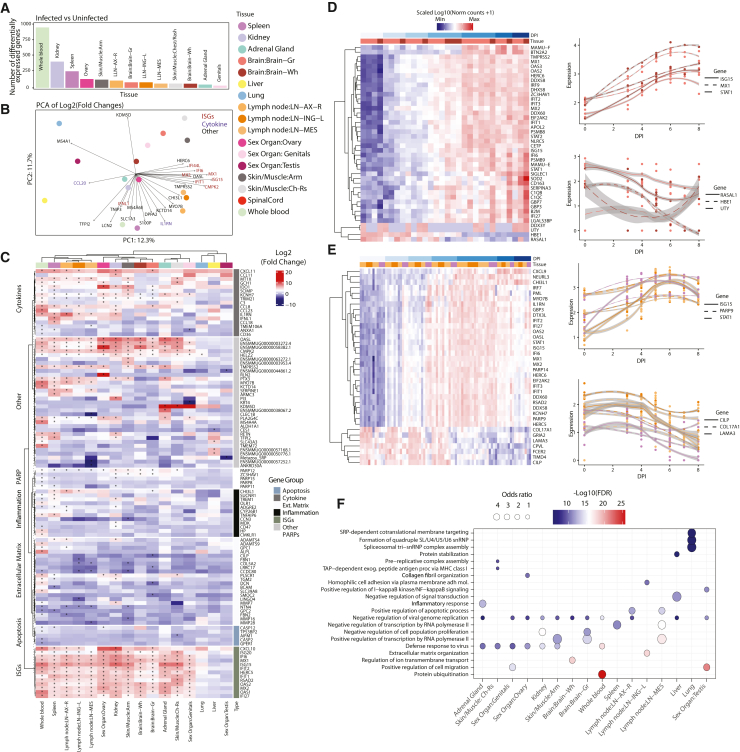
Figure 3.
Host transcriptomics across tissues and time
(A) Number of DEGs between non-infected and infected samples; tissues with more than 5 DEGs are shown in the plot.
(B) PCA of log2 fold changes of significantly DEGs between infected and uninfected samples. Top contributing genes for PC1 and PC2 are highlighted.
(C) Heatmap of fold-changes of top DEGs across tissues, stratified by meaningful gene categories; stars marks significant differential expression (FDR < 0.05).
(D) Left: heatmap of genes changing significantly across time for brain. Right: gene expression changes across time for selected genes. Colors atop plots designate gray (light red) and white matter (dark red).
(E) Same as (D) but for lymph nodes (shades of orange) and spleen (purple); colors atop plots designate tissues.
(F) Gene Ontology (GO) term analysis of genes differentially expressed (top 100 FDR < 0.01) across time as determined by ImpulseDE2. Enriched terms were determined per tissue, and the top 3 GO terms, as determined by Kolmogorov-Smirnov (KS) test, per tissue were selected for display. Colors of circles correspond to −log10(KS pval) of the enriched term within tissue, and sizes of circles correspond to odds ratio.