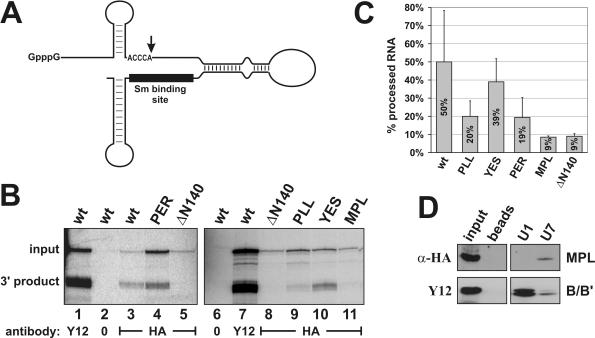
Figure 5.
Three conserved amino acid sequences in the N-terminus of Lsm11 are important for histone RNA 3′ end processing. (A) The chimeric histone–U7 RNA [12/12-U7 RNA (25)] used in (B) contains 49 nt of histone pre-mRNA upstream and 36 nt downstream of the cleavage site (vertical arrow), a connector segment of 28 and 65 nt of U7 RNA sequence. The Sm-binding site is indicated by a black bar. (B) Processing of chimeric histone–U7 RNA in Xenopus oocytes. Synthetic mRNAs encoding HA-tagged versions of mLsm11FL (wt) or of the four mutants (see Figure 3A) were injected into the cytoplasm of X.laevis oocytes. RNA encoding HA-tagged Lsm11 lacking the first 140 amino acids (ΔN140) was injected as a processing-deficient control (16). After overnight incubation to allow for translation of the recombinant proteins, the oocytes were challenged with radiolabelled, chimeric histone–U7 RNA [see (A)]. In the oocytes, this RNA gets cleaved at the histone RNA processing site, dependent upon assembly of a functional Sm/Lsm core at its Sm-binding site. Oocyte extracts were subjected to immunoprecipitation with either Y12 anti-Sm or anti-HA antibodies to enrich for total snRNPs or for particles containing HA-tagged Lsm11, respectively. The radiolabelled RNA was analysed by denaturing polyacrylamide gel electrophoresis and autoradiography. Lanes 2 and 6, HA-mLsm11FL extract (wt) precipitated with beads lacking antibody. (C) Quantitation of the ratios of 3′ processing product:total chimeric RNA determined by PhosphorImager scanning. The results represent averages of 2–3 independent determinations for each construct. Note that the MPL mutant and the N-terminal deletion show a strong defect in processing, but still associate with the chimeric RNA. The PLL and PER mutants are partly deficient in processing. (D) The MPL mutant protein is associated with U7 snRNA in mammalian cell extracts. Whole cell extract from human 293-T cells transiently transfected with the plasmid encoding HA-mLsm11MPL was incubated with biotinylated oligonucleotides complementary to the 5′ ends of either U7 or U1 snRNA and precipitated with magnetic streptavidin beads. The samples were subjected to SDS–PAGE and immunoblotted with anti-HA antibody to detect the HA-mLsm11MPL protein and with Y12 antibody to detect the Sm B/B′ protein as a precipitation control. Beads, precipitation by beads without oligonucleotide; input, 1/10 of original extract. The two panels are taken from the same gel, but an intermediate lane was excised from the picture.